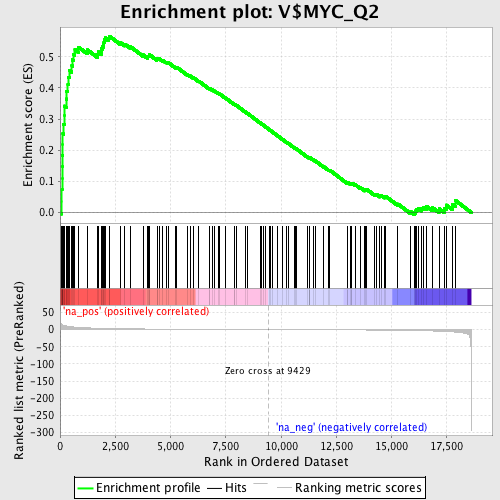
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$MYC_Q2 |
| Enrichment Score (ES) | 0.56606954 |
| Normalized Enrichment Score (NES) | 1.9453164 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0013958148 |
| FWER p-Value | 0.0080 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FKBP11 | 82 | 14.885 | 0.0344 | Yes | ||
| 2 | B3GALT6 | 83 | 14.867 | 0.0731 | Yes | ||
| 3 | ATAD3A | 91 | 14.629 | 0.1108 | Yes | ||
| 4 | ADK | 94 | 14.400 | 0.1482 | Yes | ||
| 5 | EBNA1BP2 | 101 | 14.096 | 0.1847 | Yes | ||
| 6 | LDHA | 123 | 13.322 | 0.2182 | Yes | ||
| 7 | NUDC | 126 | 13.274 | 0.2527 | Yes | ||
| 8 | AK2 | 163 | 12.258 | 0.2827 | Yes | ||
| 9 | PPRC1 | 187 | 11.752 | 0.3121 | Yes | ||
| 10 | SHMT2 | 192 | 11.577 | 0.3420 | Yes | ||
| 11 | TIMM10 | 277 | 10.400 | 0.3646 | Yes | ||
| 12 | CSDA | 298 | 10.136 | 0.3899 | Yes | ||
| 13 | PABPC4 | 354 | 9.488 | 0.4117 | Yes | ||
| 14 | AMPD2 | 382 | 9.297 | 0.4344 | Yes | ||
| 15 | PA2G4 | 417 | 9.021 | 0.4561 | Yes | ||
| 16 | COPS7A | 534 | 8.039 | 0.4708 | Yes | ||
| 17 | EIF3S10 | 555 | 7.895 | 0.4903 | Yes | ||
| 18 | ZZZ3 | 613 | 7.527 | 0.5068 | Yes | ||
| 19 | GTF2H1 | 671 | 7.198 | 0.5225 | Yes | ||
| 20 | SFXN2 | 828 | 6.502 | 0.5310 | Yes | ||
| 21 | IVNS1ABP | 1236 | 5.009 | 0.5220 | Yes | ||
| 22 | IL15RA | 1693 | 3.875 | 0.5075 | Yes | ||
| 23 | HNRPA1 | 1726 | 3.812 | 0.5157 | Yes | ||
| 24 | CENTB5 | 1869 | 3.511 | 0.5172 | Yes | ||
| 25 | CHD4 | 1881 | 3.494 | 0.5257 | Yes | ||
| 26 | BHLHB3 | 1908 | 3.447 | 0.5332 | Yes | ||
| 27 | RHEBL1 | 1956 | 3.372 | 0.5395 | Yes | ||
| 28 | EIF4B | 1981 | 3.329 | 0.5469 | Yes | ||
| 29 | ALDH3B1 | 2003 | 3.292 | 0.5543 | Yes | ||
| 30 | PFDN2 | 2044 | 3.232 | 0.5606 | Yes | ||
| 31 | ARL3 | 2212 | 2.981 | 0.5593 | Yes | ||
| 32 | TAGLN2 | 2230 | 2.945 | 0.5661 | Yes | ||
| 33 | FABP3 | 2729 | 2.411 | 0.5454 | No | ||
| 34 | TFB2M | 2935 | 2.233 | 0.5402 | No | ||
| 35 | RAB3IL1 | 3178 | 2.046 | 0.5324 | No | ||
| 36 | SYT6 | 3751 | 1.680 | 0.5059 | No | ||
| 37 | RFX5 | 3954 | 1.579 | 0.4991 | No | ||
| 38 | WEE1 | 3977 | 1.567 | 0.5020 | No | ||
| 39 | LMX1A | 4025 | 1.546 | 0.5035 | No | ||
| 40 | RORC | 4040 | 1.540 | 0.5067 | No | ||
| 41 | B3GALT2 | 4385 | 1.397 | 0.4918 | No | ||
| 42 | CFL1 | 4411 | 1.389 | 0.4940 | No | ||
| 43 | HOXC13 | 4487 | 1.355 | 0.4935 | No | ||
| 44 | STMN1 | 4636 | 1.302 | 0.4889 | No | ||
| 45 | CTBP2 | 4805 | 1.231 | 0.4830 | No | ||
| 46 | GPD1 | 4889 | 1.201 | 0.4817 | No | ||
| 47 | TIAL1 | 5227 | 1.100 | 0.4663 | No | ||
| 48 | LHX9 | 5282 | 1.086 | 0.4662 | No | ||
| 49 | SC5DL | 5779 | 0.932 | 0.4418 | No | ||
| 50 | CNNM1 | 5886 | 0.900 | 0.4384 | No | ||
| 51 | IPO7 | 6015 | 0.863 | 0.4338 | No | ||
| 52 | FCHSD2 | 6274 | 0.793 | 0.4219 | No | ||
| 53 | FXYD6 | 6750 | 0.675 | 0.3980 | No | ||
| 54 | SLC16A1 | 6767 | 0.668 | 0.3989 | No | ||
| 55 | RTN4RL2 | 6890 | 0.635 | 0.3939 | No | ||
| 56 | PITX3 | 6988 | 0.609 | 0.3903 | No | ||
| 57 | HNRPF | 7168 | 0.567 | 0.3821 | No | ||
| 58 | PLA2G4A | 7230 | 0.552 | 0.3802 | No | ||
| 59 | HOXC12 | 7480 | 0.488 | 0.3680 | No | ||
| 60 | FADS3 | 7880 | 0.386 | 0.3475 | No | ||
| 61 | KRTCAP2 | 7889 | 0.384 | 0.3480 | No | ||
| 62 | LTBR | 7993 | 0.359 | 0.3434 | No | ||
| 63 | TGFB2 | 8377 | 0.261 | 0.3234 | No | ||
| 64 | POGK | 8472 | 0.240 | 0.3189 | No | ||
| 65 | FOSL1 | 9050 | 0.097 | 0.2880 | No | ||
| 66 | POU3F1 | 9135 | 0.075 | 0.2836 | No | ||
| 67 | ATP5F1 | 9182 | 0.061 | 0.2813 | No | ||
| 68 | TESK2 | 9279 | 0.037 | 0.2762 | No | ||
| 69 | IPO13 | 9493 | -0.020 | 0.2647 | No | ||
| 70 | RPL22 | 9540 | -0.034 | 0.2623 | No | ||
| 71 | NRAS | 9596 | -0.046 | 0.2595 | No | ||
| 72 | HPCA | 9840 | -0.111 | 0.2466 | No | ||
| 73 | OPRD1 | 10062 | -0.168 | 0.2351 | No | ||
| 74 | UBXD3 | 10250 | -0.211 | 0.2256 | No | ||
| 75 | PTPRF | 10334 | -0.228 | 0.2217 | No | ||
| 76 | PAX6 | 10602 | -0.286 | 0.2080 | No | ||
| 77 | NRIP3 | 10659 | -0.297 | 0.2057 | No | ||
| 78 | USP2 | 10680 | -0.302 | 0.2055 | No | ||
| 79 | AGMAT | 11205 | -0.436 | 0.1783 | No | ||
| 80 | NKX2-3 | 11279 | -0.454 | 0.1755 | No | ||
| 81 | HMGN2 | 11287 | -0.455 | 0.1763 | No | ||
| 82 | SLC1A7 | 11292 | -0.456 | 0.1773 | No | ||
| 83 | CGN | 11457 | -0.498 | 0.1697 | No | ||
| 84 | LIN28 | 11552 | -0.520 | 0.1660 | No | ||
| 85 | COL2A1 | 11924 | -0.626 | 0.1476 | No | ||
| 86 | ATP6V0B | 12166 | -0.694 | 0.1363 | No | ||
| 87 | CTSF | 12201 | -0.703 | 0.1363 | No | ||
| 88 | TAF6L | 13014 | -0.948 | 0.0949 | No | ||
| 89 | ALX4 | 13016 | -0.949 | 0.0973 | No | ||
| 90 | SLC6A15 | 13141 | -0.995 | 0.0932 | No | ||
| 91 | SCYL1 | 13169 | -1.005 | 0.0944 | No | ||
| 92 | NR0B2 | 13207 | -1.018 | 0.0950 | No | ||
| 93 | B4GALT2 | 13365 | -1.074 | 0.0893 | No | ||
| 94 | APOA5 | 13574 | -1.159 | 0.0811 | No | ||
| 95 | TDRD1 | 13791 | -1.247 | 0.0727 | No | ||
| 96 | FOXD3 | 13823 | -1.256 | 0.0743 | No | ||
| 97 | CHRM1 | 13857 | -1.270 | 0.0758 | No | ||
| 98 | LRP8 | 14231 | -1.409 | 0.0593 | No | ||
| 99 | FXYD2 | 14333 | -1.452 | 0.0576 | No | ||
| 100 | RUSC1 | 14473 | -1.511 | 0.0541 | No | ||
| 101 | AP3M1 | 14530 | -1.538 | 0.0550 | No | ||
| 102 | TMEM24 | 14681 | -1.620 | 0.0512 | No | ||
| 103 | HPCAL4 | 14722 | -1.637 | 0.0533 | No | ||
| 104 | CBX5 | 15290 | -2.008 | 0.0278 | No | ||
| 105 | TRIM46 | 15852 | -2.479 | 0.0040 | No | ||
| 106 | CDC14A | 16035 | -2.699 | 0.0012 | No | ||
| 107 | ILK | 16089 | -2.770 | 0.0055 | No | ||
| 108 | USP15 | 16132 | -2.823 | 0.0106 | No | ||
| 109 | NIT1 | 16219 | -2.927 | 0.0136 | No | ||
| 110 | PFKFB3 | 16375 | -3.169 | 0.0135 | No | ||
| 111 | BCL9L | 16456 | -3.299 | 0.0177 | No | ||
| 112 | COMMD3 | 16590 | -3.544 | 0.0198 | No | ||
| 113 | SIRT1 | 16844 | -4.003 | 0.0165 | No | ||
| 114 | H3F3A | 17152 | -4.820 | 0.0125 | No | ||
| 115 | RRAGC | 17384 | -5.489 | 0.0143 | No | ||
| 116 | ESRRA | 17475 | -5.806 | 0.0246 | No | ||
| 117 | PSEN2 | 17754 | -6.742 | 0.0271 | No | ||
| 118 | ATF7IP | 17898 | -7.456 | 0.0388 | No |