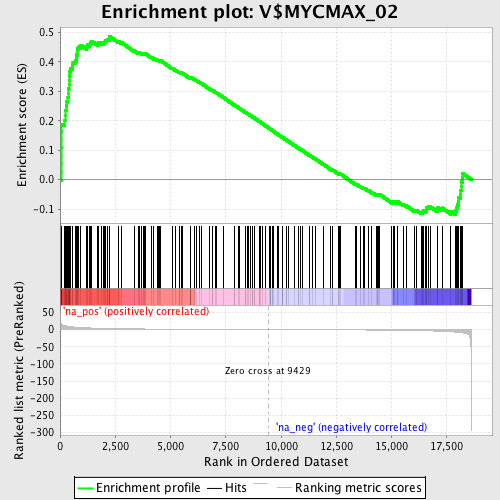
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$MYCMAX_02 |
| Enrichment Score (ES) | 0.48708403 |
| Normalized Enrichment Score (NES) | 1.7883841 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.011864333 |
| FWER p-Value | 0.129 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CIRH1A | 45 | 17.130 | 0.0260 | Yes | ||
| 2 | FBL | 47 | 16.998 | 0.0542 | Yes | ||
| 3 | RCL1 | 49 | 16.885 | 0.0821 | Yes | ||
| 4 | POLR3E | 50 | 16.879 | 0.1102 | Yes | ||
| 5 | ACY1 | 60 | 16.287 | 0.1367 | Yes | ||
| 6 | TIMM8A | 61 | 16.206 | 0.1636 | Yes | ||
| 7 | CAD | 74 | 15.227 | 0.1882 | Yes | ||
| 8 | PPRC1 | 187 | 11.752 | 0.2017 | Yes | ||
| 9 | HDGF | 225 | 11.084 | 0.2181 | Yes | ||
| 10 | DPAGT1 | 230 | 10.951 | 0.2360 | Yes | ||
| 11 | TIMM10 | 277 | 10.400 | 0.2508 | Yes | ||
| 12 | MTHFD1 | 307 | 10.020 | 0.2659 | Yes | ||
| 13 | PABPC4 | 354 | 9.488 | 0.2791 | Yes | ||
| 14 | NOLC1 | 378 | 9.329 | 0.2934 | Yes | ||
| 15 | AMPD2 | 382 | 9.297 | 0.3086 | Yes | ||
| 16 | NACA | 404 | 9.137 | 0.3227 | Yes | ||
| 17 | RPS2 | 410 | 9.090 | 0.3375 | Yes | ||
| 18 | PA2G4 | 417 | 9.021 | 0.3521 | Yes | ||
| 19 | SYNCRIP | 429 | 8.944 | 0.3664 | Yes | ||
| 20 | EXOSC5 | 472 | 8.524 | 0.3782 | Yes | ||
| 21 | MORF4L2 | 564 | 7.838 | 0.3863 | Yes | ||
| 22 | USP36 | 580 | 7.752 | 0.3984 | Yes | ||
| 23 | FKBP5 | 675 | 7.146 | 0.4051 | Yes | ||
| 24 | GPS1 | 735 | 6.849 | 0.4133 | Yes | ||
| 25 | UMPS | 751 | 6.770 | 0.4237 | Yes | ||
| 26 | POLR2H | 779 | 6.679 | 0.4334 | Yes | ||
| 27 | NR1D1 | 782 | 6.668 | 0.4443 | Yes | ||
| 28 | HSPA4 | 852 | 6.371 | 0.4512 | Yes | ||
| 29 | HNRPDL | 941 | 6.014 | 0.4564 | Yes | ||
| 30 | MAT2A | 1206 | 5.097 | 0.4505 | Yes | ||
| 31 | IVNS1ABP | 1236 | 5.009 | 0.4573 | Yes | ||
| 32 | CDK2 | 1330 | 4.747 | 0.4601 | Yes | ||
| 33 | PSME3 | 1374 | 4.598 | 0.4654 | Yes | ||
| 34 | ADD3 | 1425 | 4.464 | 0.4701 | Yes | ||
| 35 | HSPH1 | 1686 | 3.890 | 0.4625 | Yes | ||
| 36 | HNRPA1 | 1726 | 3.812 | 0.4667 | Yes | ||
| 37 | FOXF2 | 1867 | 3.516 | 0.4649 | Yes | ||
| 38 | EIF4B | 1981 | 3.329 | 0.4643 | Yes | ||
| 39 | FBXW7 | 2029 | 3.259 | 0.4672 | Yes | ||
| 40 | ANKRD17 | 2053 | 3.218 | 0.4713 | Yes | ||
| 41 | GATA4 | 2122 | 3.119 | 0.4728 | Yes | ||
| 42 | INSM1 | 2159 | 3.065 | 0.4759 | Yes | ||
| 43 | TGIF2 | 2223 | 2.960 | 0.4774 | Yes | ||
| 44 | DSCAM | 2226 | 2.953 | 0.4822 | Yes | ||
| 45 | CITED1 | 2227 | 2.951 | 0.4871 | Yes | ||
| 46 | CLCN2 | 2620 | 2.501 | 0.4700 | No | ||
| 47 | LRP5 | 2767 | 2.380 | 0.4660 | No | ||
| 48 | NPM1 | 3360 | 1.903 | 0.4371 | No | ||
| 49 | FGF14 | 3542 | 1.794 | 0.4302 | No | ||
| 50 | CHST11 | 3587 | 1.767 | 0.4308 | No | ||
| 51 | TIMP3 | 3680 | 1.720 | 0.4287 | No | ||
| 52 | SYT6 | 3751 | 1.680 | 0.4276 | No | ||
| 53 | GPR65 | 3826 | 1.640 | 0.4264 | No | ||
| 54 | PCDHA10 | 3848 | 1.628 | 0.4279 | No | ||
| 55 | HOXC4 | 4118 | 1.511 | 0.4158 | No | ||
| 56 | SEPT3 | 4248 | 1.452 | 0.4113 | No | ||
| 57 | B3GALT2 | 4385 | 1.397 | 0.4062 | No | ||
| 58 | HIF1A | 4444 | 1.376 | 0.4053 | No | ||
| 59 | TOP3A | 4510 | 1.348 | 0.4040 | No | ||
| 60 | CSMD3 | 4526 | 1.343 | 0.4055 | No | ||
| 61 | IGF1R | 5072 | 1.151 | 0.3778 | No | ||
| 62 | RUNX2 | 5229 | 1.100 | 0.3712 | No | ||
| 63 | ANGPT2 | 5394 | 1.044 | 0.3640 | No | ||
| 64 | DIABLO | 5408 | 1.040 | 0.3651 | No | ||
| 65 | SEMA7A | 5485 | 1.020 | 0.3626 | No | ||
| 66 | ATOH8 | 5550 | 0.998 | 0.3608 | No | ||
| 67 | DCT | 5884 | 0.900 | 0.3443 | No | ||
| 68 | CNNM1 | 5886 | 0.900 | 0.3457 | No | ||
| 69 | SILV | 5890 | 0.900 | 0.3470 | No | ||
| 70 | RAI14 | 5899 | 0.897 | 0.3481 | No | ||
| 71 | TYRP1 | 6076 | 0.846 | 0.3399 | No | ||
| 72 | NEUROD6 | 6165 | 0.822 | 0.3365 | No | ||
| 73 | HOXD10 | 6295 | 0.788 | 0.3308 | No | ||
| 74 | LZTS2 | 6382 | 0.769 | 0.3275 | No | ||
| 75 | PTMA | 6756 | 0.671 | 0.3083 | No | ||
| 76 | RTN4RL2 | 6890 | 0.635 | 0.3022 | No | ||
| 77 | TOP1 | 6905 | 0.631 | 0.3025 | No | ||
| 78 | CDK5R1 | 7018 | 0.604 | 0.2974 | No | ||
| 79 | EN1 | 7062 | 0.593 | 0.2961 | No | ||
| 80 | GSH1 | 7410 | 0.511 | 0.2781 | No | ||
| 81 | SOX10 | 7874 | 0.387 | 0.2536 | No | ||
| 82 | BOK | 7887 | 0.384 | 0.2536 | No | ||
| 83 | PTK7 | 8070 | 0.340 | 0.2443 | No | ||
| 84 | DBP | 8120 | 0.328 | 0.2422 | No | ||
| 85 | TGFB2 | 8377 | 0.261 | 0.2288 | No | ||
| 86 | PLAG1 | 8396 | 0.256 | 0.2282 | No | ||
| 87 | POGK | 8472 | 0.240 | 0.2245 | No | ||
| 88 | LRRC2 | 8515 | 0.230 | 0.2226 | No | ||
| 89 | HOXB5 | 8599 | 0.209 | 0.2185 | No | ||
| 90 | CALB2 | 8705 | 0.182 | 0.2131 | No | ||
| 91 | XPO1 | 8722 | 0.178 | 0.2125 | No | ||
| 92 | UBE2B | 8785 | 0.165 | 0.2094 | No | ||
| 93 | FGF20 | 8799 | 0.161 | 0.2090 | No | ||
| 94 | NEUROD2 | 9002 | 0.111 | 0.1982 | No | ||
| 95 | TYRO3 | 9032 | 0.104 | 0.1968 | No | ||
| 96 | MAX | 9067 | 0.094 | 0.1951 | No | ||
| 97 | SMCR8 | 9151 | 0.068 | 0.1907 | No | ||
| 98 | TESK2 | 9279 | 0.037 | 0.1839 | No | ||
| 99 | IPO13 | 9493 | -0.020 | 0.1724 | No | ||
| 100 | NPHS1 | 9514 | -0.026 | 0.1714 | No | ||
| 101 | SLC12A4 | 9531 | -0.032 | 0.1705 | No | ||
| 102 | DLX1 | 9542 | -0.034 | 0.1701 | No | ||
| 103 | ODZ1 | 9595 | -0.046 | 0.1673 | No | ||
| 104 | ENPP2 | 9611 | -0.050 | 0.1666 | No | ||
| 105 | RNF12 | 9670 | -0.065 | 0.1636 | No | ||
| 106 | HPCA | 9840 | -0.111 | 0.1546 | No | ||
| 107 | TNFRSF21 | 9890 | -0.125 | 0.1521 | No | ||
| 108 | OPRD1 | 10062 | -0.168 | 0.1431 | No | ||
| 109 | RASGEF1B | 10067 | -0.170 | 0.1432 | No | ||
| 110 | ADAMTS17 | 10074 | -0.172 | 0.1432 | No | ||
| 111 | C1QTNF5 | 10241 | -0.209 | 0.1345 | No | ||
| 112 | UBXD3 | 10250 | -0.211 | 0.1344 | No | ||
| 113 | NR5A1 | 10341 | -0.229 | 0.1299 | No | ||
| 114 | SOX12 | 10623 | -0.289 | 0.1152 | No | ||
| 115 | HOXB7 | 10776 | -0.327 | 0.1075 | No | ||
| 116 | PRKCH | 10782 | -0.328 | 0.1077 | No | ||
| 117 | APLN | 10870 | -0.350 | 0.1036 | No | ||
| 118 | MKRN3 | 10966 | -0.377 | 0.0991 | No | ||
| 119 | GPR17 | 11289 | -0.455 | 0.0824 | No | ||
| 120 | SLC1A7 | 11292 | -0.456 | 0.0830 | No | ||
| 121 | SMAD2 | 11415 | -0.487 | 0.0772 | No | ||
| 122 | LIN28 | 11552 | -0.520 | 0.0707 | No | ||
| 123 | COL2A1 | 11924 | -0.626 | 0.0516 | No | ||
| 124 | SATB2 | 12217 | -0.707 | 0.0370 | No | ||
| 125 | CD2 | 12317 | -0.741 | 0.0328 | No | ||
| 126 | RNF128 | 12334 | -0.746 | 0.0332 | No | ||
| 127 | GALNT4 | 12603 | -0.831 | 0.0200 | No | ||
| 128 | GPC3 | 12612 | -0.833 | 0.0210 | No | ||
| 129 | FZD6 | 12627 | -0.837 | 0.0216 | No | ||
| 130 | OGN | 12712 | -0.864 | 0.0185 | No | ||
| 131 | IRS4 | 13356 | -1.072 | -0.0146 | No | ||
| 132 | HOXA11 | 13400 | -1.088 | -0.0151 | No | ||
| 133 | BACH2 | 13573 | -1.159 | -0.0225 | No | ||
| 134 | VNN1 | 13726 | -1.223 | -0.0287 | No | ||
| 135 | MAN2A2 | 13768 | -1.237 | -0.0289 | No | ||
| 136 | TYR | 13978 | -1.313 | -0.0381 | No | ||
| 137 | LEF1 | 14115 | -1.368 | -0.0432 | No | ||
| 138 | WDR17 | 14314 | -1.444 | -0.0515 | No | ||
| 139 | BCOR | 14348 | -1.458 | -0.0509 | No | ||
| 140 | CBARA1 | 14356 | -1.461 | -0.0488 | No | ||
| 141 | H2AFZ | 14415 | -1.488 | -0.0495 | No | ||
| 142 | HOXA3 | 14446 | -1.500 | -0.0486 | No | ||
| 143 | LPXN | 15017 | -1.808 | -0.0765 | No | ||
| 144 | ARF6 | 15073 | -1.843 | -0.0765 | No | ||
| 145 | RAB35 | 15076 | -1.846 | -0.0735 | No | ||
| 146 | BMP2K | 15135 | -1.887 | -0.0735 | No | ||
| 147 | H3F3B | 15288 | -2.007 | -0.0784 | No | ||
| 148 | CBX5 | 15290 | -2.008 | -0.0751 | No | ||
| 149 | JMJD1C | 15536 | -2.190 | -0.0848 | No | ||
| 150 | SGCD | 15684 | -2.322 | -0.0889 | No | ||
| 151 | BCKDHA | 16060 | -2.729 | -0.1047 | No | ||
| 152 | USP15 | 16132 | -2.823 | -0.1039 | No | ||
| 153 | TCF4 | 16340 | -3.121 | -0.1099 | No | ||
| 154 | EFNB1 | 16407 | -3.227 | -0.1081 | No | ||
| 155 | BCL9L | 16456 | -3.299 | -0.1053 | No | ||
| 156 | CDKL5 | 16550 | -3.473 | -0.1046 | No | ||
| 157 | COMMD3 | 16590 | -3.544 | -0.1008 | No | ||
| 158 | BACH1 | 16593 | -3.550 | -0.0950 | No | ||
| 159 | HOXA7 | 16657 | -3.671 | -0.0923 | No | ||
| 160 | PPM1A | 16780 | -3.890 | -0.0925 | No | ||
| 161 | WDR23 | 17086 | -4.638 | -0.1013 | No | ||
| 162 | NOTCH1 | 17098 | -4.678 | -0.0941 | No | ||
| 163 | MBNL1 | 17291 | -5.195 | -0.0959 | No | ||
| 164 | PPP1R9B | 17682 | -6.443 | -0.1064 | No | ||
| 165 | ATF7IP | 17898 | -7.456 | -0.1057 | No | ||
| 166 | STAT3 | 17952 | -7.686 | -0.0958 | No | ||
| 167 | ETV1 | 17982 | -7.808 | -0.0844 | No | ||
| 168 | CDKN2C | 18019 | -8.014 | -0.0730 | No | ||
| 169 | LEMD2 | 18033 | -8.107 | -0.0603 | No | ||
| 170 | ARRDC3 | 18128 | -8.916 | -0.0506 | No | ||
| 171 | KBTBD2 | 18139 | -8.997 | -0.0362 | No | ||
| 172 | IHPK1 | 18152 | -9.044 | -0.0218 | No | ||
| 173 | ISGF3G | 18156 | -9.078 | -0.0069 | No | ||
| 174 | CSK | 18203 | -9.505 | 0.0064 | No | ||
| 175 | TP53 | 18216 | -9.624 | 0.0217 | No |
