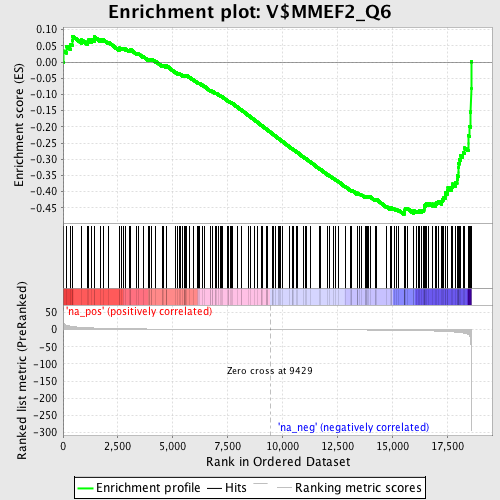
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$MMEF2_Q6 |
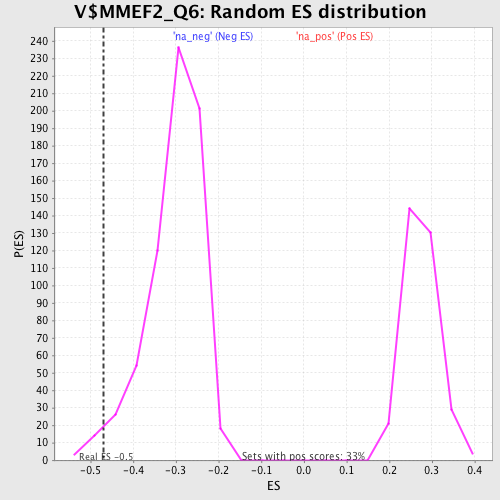
| Enrichment Score (ES) | -0.4704797 |
| Normalized Enrichment Score (NES) | -1.5457801 |
| Nominal p-value | 0.023809524 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PDLIM1 | 23 | 21.160 | 0.0343 | No | ||
| 2 | CRTAP | 144 | 12.799 | 0.0493 | No | ||
| 3 | BZW2 | 341 | 9.694 | 0.0550 | No | ||
| 4 | NEDD4 | 418 | 9.012 | 0.0660 | No | ||
| 5 | NANOS1 | 433 | 8.880 | 0.0802 | No | ||
| 6 | GYPC | 845 | 6.412 | 0.0686 | No | ||
| 7 | DNAJA2 | 1129 | 5.355 | 0.0623 | No | ||
| 8 | RBM8A | 1174 | 5.199 | 0.0686 | No | ||
| 9 | LUC7L | 1311 | 4.817 | 0.0693 | No | ||
| 10 | ADD3 | 1425 | 4.464 | 0.0707 | No | ||
| 11 | LUZP1 | 1429 | 4.452 | 0.0780 | No | ||
| 12 | GCAT | 1687 | 3.890 | 0.0706 | No | ||
| 13 | ELMO3 | 1831 | 3.593 | 0.0689 | No | ||
| 14 | COX7A2 | 2059 | 3.208 | 0.0620 | No | ||
| 15 | ACTG2 | 2555 | 2.565 | 0.0394 | No | ||
| 16 | ANKMY2 | 2556 | 2.565 | 0.0437 | No | ||
| 17 | DUSP14 | 2641 | 2.486 | 0.0434 | No | ||
| 18 | MAFA | 2743 | 2.401 | 0.0419 | No | ||
| 19 | DMD | 2839 | 2.319 | 0.0407 | No | ||
| 20 | TOMM70A | 3006 | 2.170 | 0.0353 | No | ||
| 21 | SIN3A | 3059 | 2.132 | 0.0361 | No | ||
| 22 | PACSIN3 | 3086 | 2.114 | 0.0382 | No | ||
| 23 | PPP1R16B | 3350 | 1.913 | 0.0272 | No | ||
| 24 | SALL1 | 3440 | 1.852 | 0.0254 | No | ||
| 25 | ELAVL4 | 3646 | 1.739 | 0.0172 | No | ||
| 26 | GRIK1 | 3901 | 1.601 | 0.0061 | No | ||
| 27 | NRAP | 3928 | 1.591 | 0.0074 | No | ||
| 28 | HAS2 | 3958 | 1.577 | 0.0085 | No | ||
| 29 | LMX1A | 4025 | 1.546 | 0.0075 | No | ||
| 30 | TBX4 | 4047 | 1.535 | 0.0089 | No | ||
| 31 | HNRPL | 4226 | 1.461 | 0.0017 | No | ||
| 32 | POU3F2 | 4523 | 1.343 | -0.0121 | No | ||
| 33 | KPNA3 | 4550 | 1.334 | -0.0112 | No | ||
| 34 | UBE3A | 4564 | 1.330 | -0.0097 | No | ||
| 35 | SEMA3A | 4705 | 1.273 | -0.0152 | No | ||
| 36 | DKK1 | 4709 | 1.272 | -0.0132 | No | ||
| 37 | BNC2 | 4711 | 1.271 | -0.0111 | No | ||
| 38 | MYH6 | 5130 | 1.131 | -0.0319 | No | ||
| 39 | RUNX2 | 5229 | 1.100 | -0.0353 | No | ||
| 40 | LHX9 | 5282 | 1.086 | -0.0363 | No | ||
| 41 | NHLH1 | 5340 | 1.070 | -0.0376 | No | ||
| 42 | ACCN1 | 5458 | 1.026 | -0.0423 | No | ||
| 43 | MYOZ2 | 5525 | 1.007 | -0.0441 | No | ||
| 44 | KBTBD5 | 5528 | 1.006 | -0.0426 | No | ||
| 45 | TBR1 | 5541 | 1.001 | -0.0415 | No | ||
| 46 | ADAM11 | 5579 | 0.990 | -0.0419 | No | ||
| 47 | SLC26A9 | 5589 | 0.987 | -0.0407 | No | ||
| 48 | HOXA10 | 5612 | 0.981 | -0.0403 | No | ||
| 49 | GABRA1 | 5755 | 0.939 | -0.0464 | No | ||
| 50 | DCTN1 | 5928 | 0.891 | -0.0542 | No | ||
| 51 | CACNA2D3 | 6125 | 0.832 | -0.0634 | No | ||
| 52 | BHLHB5 | 6176 | 0.820 | -0.0648 | No | ||
| 53 | PFTK1 | 6215 | 0.809 | -0.0655 | No | ||
| 54 | FHOD1 | 6335 | 0.778 | -0.0706 | No | ||
| 55 | CYP26B1 | 6452 | 0.750 | -0.0757 | No | ||
| 56 | SMPX | 6725 | 0.681 | -0.0893 | No | ||
| 57 | CALCR | 6734 | 0.678 | -0.0886 | No | ||
| 58 | HOXD4 | 6808 | 0.658 | -0.0914 | No | ||
| 59 | DNAJA4 | 6822 | 0.654 | -0.0910 | No | ||
| 60 | WFDC1 | 6952 | 0.620 | -0.0970 | No | ||
| 61 | SH3BGRL3 | 6967 | 0.614 | -0.0967 | No | ||
| 62 | TCEA3 | 6985 | 0.610 | -0.0966 | No | ||
| 63 | RALY | 7079 | 0.591 | -0.1007 | No | ||
| 64 | USP13 | 7170 | 0.567 | -0.1046 | No | ||
| 65 | TACR1 | 7215 | 0.555 | -0.1061 | No | ||
| 66 | RIMS2 | 7245 | 0.550 | -0.1067 | No | ||
| 67 | GPR153 | 7486 | 0.487 | -0.1189 | No | ||
| 68 | VANGL1 | 7532 | 0.476 | -0.1205 | No | ||
| 69 | FOXA2 | 7558 | 0.471 | -0.1211 | No | ||
| 70 | SSH3 | 7633 | 0.448 | -0.1244 | No | ||
| 71 | COL11A2 | 7657 | 0.444 | -0.1249 | No | ||
| 72 | CRHBP | 7727 | 0.423 | -0.1279 | No | ||
| 73 | ACTA1 | 7930 | 0.374 | -0.1382 | No | ||
| 74 | MYL1 | 8113 | 0.330 | -0.1476 | No | ||
| 75 | SV2A | 8152 | 0.317 | -0.1491 | No | ||
| 76 | PRX | 8428 | 0.251 | -0.1636 | No | ||
| 77 | ELAVL2 | 8546 | 0.220 | -0.1696 | No | ||
| 78 | HOXD8 | 8717 | 0.179 | -0.1785 | No | ||
| 79 | NOG | 8841 | 0.153 | -0.1849 | No | ||
| 80 | MYL2 | 9026 | 0.105 | -0.1947 | No | ||
| 81 | TYRO3 | 9032 | 0.104 | -0.1948 | No | ||
| 82 | HOXC10 | 9098 | 0.086 | -0.1982 | No | ||
| 83 | CKM | 9260 | 0.042 | -0.2069 | No | ||
| 84 | BAI3 | 9280 | 0.037 | -0.2078 | No | ||
| 85 | CLASP1 | 9312 | 0.029 | -0.2095 | No | ||
| 86 | OLIG3 | 9332 | 0.023 | -0.2104 | No | ||
| 87 | HSPB1 | 9537 | -0.034 | -0.2215 | No | ||
| 88 | GSC | 9545 | -0.035 | -0.2218 | No | ||
| 89 | MAS1 | 9574 | -0.040 | -0.2232 | No | ||
| 90 | KIRREL3 | 9697 | -0.071 | -0.2297 | No | ||
| 91 | LYN | 9815 | -0.105 | -0.2359 | No | ||
| 92 | LRRTM1 | 9872 | -0.119 | -0.2387 | No | ||
| 93 | SIX3 | 9904 | -0.130 | -0.2402 | No | ||
| 94 | TOPBP1 | 9989 | -0.149 | -0.2445 | No | ||
| 95 | CLDN14 | 10313 | -0.224 | -0.2617 | No | ||
| 96 | ARHGAP12 | 10444 | -0.249 | -0.2683 | No | ||
| 97 | SLC12A5 | 10450 | -0.250 | -0.2681 | No | ||
| 98 | HTR2B | 10492 | -0.258 | -0.2699 | No | ||
| 99 | P2RX5 | 10636 | -0.292 | -0.2772 | No | ||
| 100 | USP2 | 10680 | -0.302 | -0.2790 | No | ||
| 101 | SOX3 | 10960 | -0.375 | -0.2935 | No | ||
| 102 | RCOR1 | 11062 | -0.399 | -0.2984 | No | ||
| 103 | CHN2 | 11088 | -0.404 | -0.2990 | No | ||
| 104 | HMGN2 | 11287 | -0.455 | -0.3090 | No | ||
| 105 | FLT1 | 11694 | -0.561 | -0.3301 | No | ||
| 106 | CAPN1 | 11715 | -0.566 | -0.3302 | No | ||
| 107 | NR2F1 | 12067 | -0.667 | -0.3481 | No | ||
| 108 | RTN1 | 12130 | -0.684 | -0.3504 | No | ||
| 109 | PRKCQ | 12320 | -0.741 | -0.3594 | No | ||
| 110 | MITF | 12395 | -0.766 | -0.3621 | No | ||
| 111 | ADAMTS9 | 12538 | -0.811 | -0.3684 | No | ||
| 112 | LPHN2 | 12850 | -0.903 | -0.3838 | No | ||
| 113 | HOXA4 | 13107 | -0.980 | -0.3960 | No | ||
| 114 | TWIST2 | 13154 | -0.999 | -0.3969 | No | ||
| 115 | HSPB7 | 13157 | -1.001 | -0.3953 | No | ||
| 116 | HOXA11 | 13400 | -1.088 | -0.4066 | No | ||
| 117 | HOXB6 | 13416 | -1.093 | -0.4056 | No | ||
| 118 | AMBN | 13423 | -1.096 | -0.4040 | No | ||
| 119 | ARMET | 13530 | -1.140 | -0.4079 | No | ||
| 120 | CLDN23 | 13610 | -1.176 | -0.4102 | No | ||
| 121 | SLC38A3 | 13774 | -1.239 | -0.4170 | No | ||
| 122 | INPPL1 | 13790 | -1.247 | -0.4157 | No | ||
| 123 | FOXD3 | 13823 | -1.256 | -0.4153 | No | ||
| 124 | FOXP2 | 13833 | -1.260 | -0.4137 | No | ||
| 125 | NRP2 | 13872 | -1.274 | -0.4136 | No | ||
| 126 | CACNB3 | 13935 | -1.297 | -0.4148 | No | ||
| 127 | MAB21L2 | 13997 | -1.319 | -0.4159 | No | ||
| 128 | HOXB8 | 14232 | -1.409 | -0.4262 | No | ||
| 129 | STAG2 | 14238 | -1.413 | -0.4241 | No | ||
| 130 | CASQ1 | 14302 | -1.438 | -0.4251 | No | ||
| 131 | NLK | 14717 | -1.635 | -0.4448 | No | ||
| 132 | PRDM13 | 14923 | -1.753 | -0.4530 | No | ||
| 133 | FLRT3 | 14952 | -1.769 | -0.4515 | No | ||
| 134 | PRRX1 | 14979 | -1.791 | -0.4499 | No | ||
| 135 | TLE3 | 15101 | -1.862 | -0.4534 | No | ||
| 136 | TNNC1 | 15183 | -1.920 | -0.4545 | No | ||
| 137 | ATP6V0C | 15286 | -2.005 | -0.4567 | No | ||
| 138 | POU2F1 | 15541 | -2.198 | -0.4668 | Yes | ||
| 139 | MYO15A | 15546 | -2.202 | -0.4633 | Yes | ||
| 140 | MYO3B | 15547 | -2.202 | -0.4596 | Yes | ||
| 141 | HIVEP3 | 15570 | -2.221 | -0.4571 | Yes | ||
| 142 | ESR1 | 15580 | -2.232 | -0.4538 | Yes | ||
| 143 | NFIL3 | 15606 | -2.259 | -0.4514 | Yes | ||
| 144 | NOX3 | 15703 | -2.344 | -0.4526 | Yes | ||
| 145 | FGF13 | 15966 | -2.612 | -0.4625 | Yes | ||
| 146 | MXI1 | 15985 | -2.644 | -0.4590 | Yes | ||
| 147 | ZDHHC8 | 16096 | -2.774 | -0.4603 | Yes | ||
| 148 | FGF9 | 16177 | -2.879 | -0.4598 | Yes | ||
| 149 | DHRS3 | 16241 | -2.954 | -0.4583 | Yes | ||
| 150 | CFL2 | 16345 | -3.128 | -0.4586 | Yes | ||
| 151 | MYB | 16420 | -3.240 | -0.4572 | Yes | ||
| 152 | EHD1 | 16450 | -3.290 | -0.4532 | Yes | ||
| 153 | BCL9L | 16456 | -3.299 | -0.4479 | Yes | ||
| 154 | JUNB | 16464 | -3.310 | -0.4427 | Yes | ||
| 155 | HAPLN1 | 16516 | -3.409 | -0.4398 | Yes | ||
| 156 | CDKL5 | 16550 | -3.473 | -0.4357 | Yes | ||
| 157 | ABCB4 | 16667 | -3.679 | -0.4358 | Yes | ||
| 158 | DLGAP4 | 16829 | -3.977 | -0.4379 | Yes | ||
| 159 | CSNK1E | 16961 | -4.282 | -0.4378 | Yes | ||
| 160 | GNB4 | 17014 | -4.423 | -0.4332 | Yes | ||
| 161 | PRKAG1 | 17100 | -4.681 | -0.4299 | Yes | ||
| 162 | ASPH | 17262 | -5.128 | -0.4301 | Yes | ||
| 163 | RFX3 | 17295 | -5.204 | -0.4230 | Yes | ||
| 164 | B4GALT1 | 17352 | -5.391 | -0.4170 | Yes | ||
| 165 | HIST1H1B | 17414 | -5.559 | -0.4110 | Yes | ||
| 166 | DNAJB5 | 17432 | -5.603 | -0.4025 | Yes | ||
| 167 | ARL6IP2 | 17528 | -6.024 | -0.3975 | Yes | ||
| 168 | KLF2 | 17539 | -6.041 | -0.3879 | Yes | ||
| 169 | HIBADH | 17706 | -6.554 | -0.3859 | Yes | ||
| 170 | RHOA | 17735 | -6.687 | -0.3762 | Yes | ||
| 171 | EYA1 | 17879 | -7.339 | -0.3716 | Yes | ||
| 172 | SDC1 | 17954 | -7.690 | -0.3627 | Yes | ||
| 173 | SSPN | 17972 | -7.775 | -0.3506 | Yes | ||
| 174 | INSIG2 | 18013 | -7.978 | -0.3393 | Yes | ||
| 175 | MBNL2 | 18014 | -7.993 | -0.3259 | Yes | ||
| 176 | RHOBTB1 | 18022 | -8.032 | -0.3128 | Yes | ||
| 177 | TCTA | 18060 | -8.301 | -0.3008 | Yes | ||
| 178 | PIP5K2C | 18120 | -8.794 | -0.2893 | Yes | ||
| 179 | CDC42EP3 | 18225 | -9.787 | -0.2785 | Yes | ||
| 180 | EHD4 | 18306 | -10.611 | -0.2650 | Yes | ||
| 181 | BCL6 | 18470 | -13.961 | -0.2503 | Yes | ||
| 182 | LMCD1 | 18471 | -14.033 | -0.2268 | Yes | ||
| 183 | SOX5 | 18531 | -18.503 | -0.1989 | Yes | ||
| 184 | AICDA | 18571 | -28.054 | -0.1538 | Yes | ||
| 185 | LRRN3 | 18590 | -43.284 | -0.0821 | Yes | ||
| 186 | BHLHB2 | 18597 | -49.644 | 0.0010 | Yes |