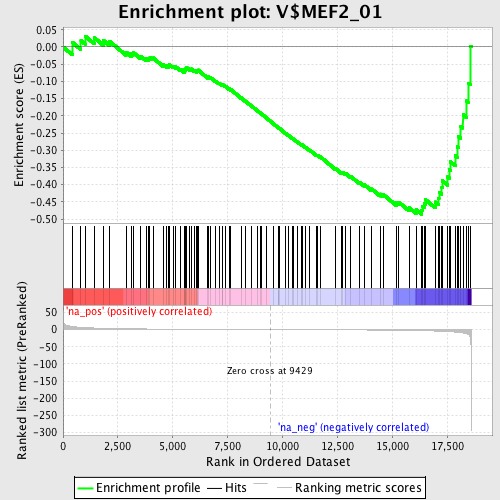
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$MEF2_01 |
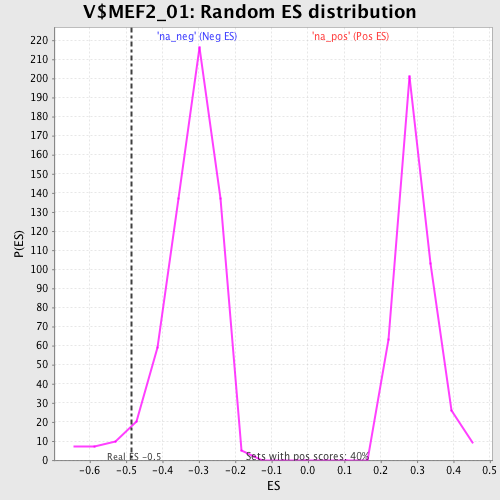
| Enrichment Score (ES) | -0.48650908 |
| Normalized Enrichment Score (NES) | -1.4969244 |
| Nominal p-value | 0.04682274 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | NEDD4 | 418 | 9.012 | 0.0136 | No | ||
| 2 | ANP32A | 813 | 6.568 | 0.0186 | No | ||
| 3 | GTF3C3 | 1005 | 5.800 | 0.0316 | No | ||
| 4 | LUZP1 | 1429 | 4.452 | 0.0266 | No | ||
| 5 | ELMO3 | 1831 | 3.593 | 0.0193 | No | ||
| 6 | GATA4 | 2122 | 3.119 | 0.0162 | No | ||
| 7 | TTYH2 | 2869 | 2.288 | -0.0149 | No | ||
| 8 | NFIX | 3105 | 2.101 | -0.0192 | No | ||
| 9 | GNAS | 3188 | 2.039 | -0.0155 | No | ||
| 10 | MYO9A | 3530 | 1.803 | -0.0266 | No | ||
| 11 | COL13A1 | 3777 | 1.668 | -0.0332 | No | ||
| 12 | GRIK1 | 3901 | 1.601 | -0.0335 | No | ||
| 13 | HAS2 | 3958 | 1.577 | -0.0302 | No | ||
| 14 | FBXO40 | 4100 | 1.516 | -0.0317 | No | ||
| 15 | UBE3A | 4564 | 1.330 | -0.0514 | No | ||
| 16 | DKK1 | 4709 | 1.272 | -0.0540 | No | ||
| 17 | AKAP12 | 4809 | 1.229 | -0.0545 | No | ||
| 18 | IGJ | 4829 | 1.221 | -0.0506 | No | ||
| 19 | MLF1 | 5017 | 1.164 | -0.0560 | No | ||
| 20 | MYH6 | 5130 | 1.131 | -0.0575 | No | ||
| 21 | SOX2 | 5352 | 1.065 | -0.0652 | No | ||
| 22 | ITGB6 | 5521 | 1.008 | -0.0702 | No | ||
| 23 | MYOZ2 | 5525 | 1.007 | -0.0664 | No | ||
| 24 | SLC26A9 | 5589 | 0.987 | -0.0658 | No | ||
| 25 | ANK3 | 5594 | 0.986 | -0.0621 | No | ||
| 26 | HOXA10 | 5612 | 0.981 | -0.0590 | No | ||
| 27 | TSC1 | 5773 | 0.933 | -0.0639 | No | ||
| 28 | RAB20 | 5838 | 0.913 | -0.0637 | No | ||
| 29 | GRAP2 | 5998 | 0.867 | -0.0688 | No | ||
| 30 | PHOSPHO1 | 6092 | 0.841 | -0.0705 | No | ||
| 31 | CACNA2D3 | 6125 | 0.832 | -0.0689 | No | ||
| 32 | DMP1 | 6148 | 0.826 | -0.0668 | No | ||
| 33 | KCNJ9 | 6598 | 0.711 | -0.0882 | No | ||
| 34 | WNT8B | 6608 | 0.708 | -0.0858 | No | ||
| 35 | SMPX | 6725 | 0.681 | -0.0893 | No | ||
| 36 | WFDC1 | 6952 | 0.620 | -0.0991 | No | ||
| 37 | TPM2 | 7110 | 0.581 | -0.1052 | No | ||
| 38 | RIMS2 | 7245 | 0.550 | -0.1102 | No | ||
| 39 | TRDN | 7266 | 0.545 | -0.1091 | No | ||
| 40 | ITGA7 | 7388 | 0.516 | -0.1136 | No | ||
| 41 | KCNN1 | 7567 | 0.469 | -0.1213 | No | ||
| 42 | MSX2 | 7650 | 0.446 | -0.1240 | No | ||
| 43 | MYL1 | 8113 | 0.330 | -0.1476 | No | ||
| 44 | SPIC | 8304 | 0.277 | -0.1567 | No | ||
| 45 | ZIC4 | 8600 | 0.208 | -0.1718 | No | ||
| 46 | ZFPM2 | 8855 | 0.148 | -0.1850 | No | ||
| 47 | FOS | 9018 | 0.107 | -0.1933 | No | ||
| 48 | MYL2 | 9026 | 0.105 | -0.1932 | No | ||
| 49 | CKM | 9260 | 0.042 | -0.2057 | No | ||
| 50 | MAS1 | 9574 | -0.040 | -0.2224 | No | ||
| 51 | PDGFRA | 9795 | -0.098 | -0.2339 | No | ||
| 52 | LYN | 9815 | -0.105 | -0.2345 | No | ||
| 53 | JPH2 | 9868 | -0.118 | -0.2368 | No | ||
| 54 | IL17B | 10129 | -0.184 | -0.2501 | No | ||
| 55 | UBXD3 | 10250 | -0.211 | -0.2558 | No | ||
| 56 | SLC12A5 | 10450 | -0.250 | -0.2655 | No | ||
| 57 | DLL4 | 10514 | -0.263 | -0.2679 | No | ||
| 58 | CKMT2 | 10696 | -0.308 | -0.2764 | No | ||
| 59 | FGF12 | 10842 | -0.343 | -0.2829 | No | ||
| 60 | EPHB1 | 10895 | -0.356 | -0.2842 | No | ||
| 61 | TNNI3K | 11057 | -0.398 | -0.2913 | No | ||
| 62 | AGTPBP1 | 11235 | -0.444 | -0.2991 | No | ||
| 63 | EDG1 | 11553 | -0.521 | -0.3141 | No | ||
| 64 | CNGA1 | 11589 | -0.532 | -0.3139 | No | ||
| 65 | CAPN1 | 11715 | -0.566 | -0.3184 | No | ||
| 66 | CASQ2 | 12394 | -0.766 | -0.3519 | No | ||
| 67 | ESRRG | 12675 | -0.851 | -0.3636 | No | ||
| 68 | TIMP2 | 12743 | -0.872 | -0.3637 | No | ||
| 69 | DPYSL3 | 12854 | -0.904 | -0.3661 | No | ||
| 70 | BMP7 | 13120 | -0.985 | -0.3764 | No | ||
| 71 | GPC4 | 13517 | -1.135 | -0.3933 | No | ||
| 72 | SYNPO2L | 13721 | -1.219 | -0.3993 | No | ||
| 73 | SESN3 | 14061 | -1.345 | -0.4122 | No | ||
| 74 | HOXA3 | 14446 | -1.500 | -0.4270 | No | ||
| 75 | ASCL2 | 14599 | -1.567 | -0.4289 | No | ||
| 76 | TNNC1 | 15183 | -1.920 | -0.4527 | No | ||
| 77 | SLC7A9 | 15291 | -2.009 | -0.4504 | No | ||
| 78 | SYT10 | 15770 | -2.404 | -0.4666 | No | ||
| 79 | USP47 | 16090 | -2.772 | -0.4727 | No | ||
| 80 | TRIM8 | 16347 | -3.129 | -0.4740 | Yes | ||
| 81 | SMARCA1 | 16368 | -3.160 | -0.4624 | Yes | ||
| 82 | CDC25B | 16457 | -3.301 | -0.4539 | Yes | ||
| 83 | HAPLN1 | 16516 | -3.409 | -0.4434 | Yes | ||
| 84 | SH3BGRL | 16960 | -4.275 | -0.4501 | Yes | ||
| 85 | PRKAG1 | 17100 | -4.681 | -0.4389 | Yes | ||
| 86 | JUN | 17135 | -4.762 | -0.4216 | Yes | ||
| 87 | ASPH | 17262 | -5.128 | -0.4079 | Yes | ||
| 88 | HDAC9 | 17284 | -5.180 | -0.3882 | Yes | ||
| 89 | ARL6IP2 | 17528 | -6.024 | -0.3772 | Yes | ||
| 90 | SMAD1 | 17599 | -6.237 | -0.3560 | Yes | ||
| 91 | HOXB4 | 17648 | -6.367 | -0.3330 | Yes | ||
| 92 | EYA1 | 17879 | -7.339 | -0.3160 | Yes | ||
| 93 | SSPN | 17972 | -7.775 | -0.2898 | Yes | ||
| 94 | MBNL2 | 18014 | -7.993 | -0.2600 | Yes | ||
| 95 | ARRDC3 | 18128 | -8.916 | -0.2303 | Yes | ||
| 96 | CDC42EP3 | 18225 | -9.787 | -0.1963 | Yes | ||
| 97 | MEF2C | 18390 | -12.088 | -0.1567 | Yes | ||
| 98 | LMCD1 | 18471 | -14.033 | -0.1047 | Yes | ||
| 99 | AICDA | 18571 | -28.054 | 0.0024 | Yes |