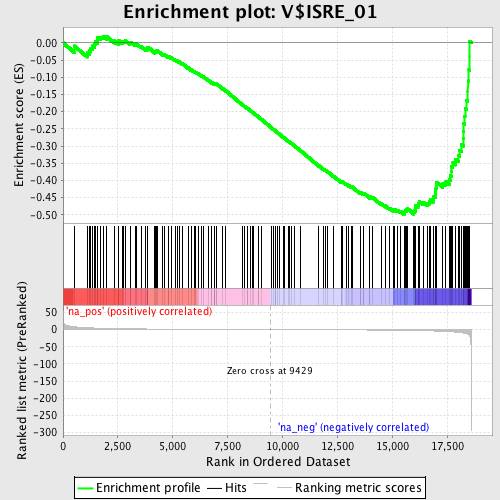
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$ISRE_01 |
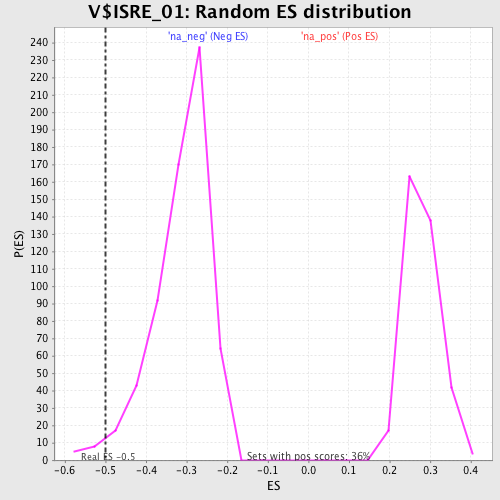
| Enrichment Score (ES) | -0.49882665 |
| Normalized Enrichment Score (NES) | -1.5877798 |
| Nominal p-value | 0.022012578 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ARHGEF6 | 505 | 8.312 | -0.0089 | No | ||
| 2 | BST2 | 1100 | 5.448 | -0.0289 | No | ||
| 3 | ANAPC1 | 1205 | 5.098 | -0.0232 | No | ||
| 4 | KCNN3 | 1266 | 4.918 | -0.0155 | No | ||
| 5 | PGK1 | 1338 | 4.727 | -0.0089 | No | ||
| 6 | ADD3 | 1425 | 4.464 | -0.0036 | No | ||
| 7 | PIK3R3 | 1472 | 4.347 | 0.0036 | No | ||
| 8 | SERHL | 1553 | 4.151 | 0.0085 | No | ||
| 9 | LY86 | 1582 | 4.102 | 0.0161 | No | ||
| 10 | MOV10 | 1723 | 3.816 | 0.0170 | No | ||
| 11 | E2F3 | 1822 | 3.614 | 0.0197 | No | ||
| 12 | ANGPTL4 | 1988 | 3.319 | 0.0181 | No | ||
| 13 | CRK | 2332 | 2.817 | 0.0058 | No | ||
| 14 | RRBP1 | 2520 | 2.594 | 0.0015 | No | ||
| 15 | HEPH | 2538 | 2.584 | 0.0063 | No | ||
| 16 | SLC25A28 | 2704 | 2.436 | 0.0028 | No | ||
| 17 | PSMA5 | 2755 | 2.386 | 0.0054 | No | ||
| 18 | FYCO1 | 2833 | 2.325 | 0.0064 | No | ||
| 19 | IL4 | 3049 | 2.137 | -0.0005 | No | ||
| 20 | FMR1 | 3089 | 2.112 | 0.0021 | No | ||
| 21 | AIF1 | 3287 | 1.962 | -0.0042 | No | ||
| 22 | EPN1 | 3345 | 1.914 | -0.0031 | No | ||
| 23 | PSMA3 | 3556 | 1.783 | -0.0105 | No | ||
| 24 | TLR7 | 3761 | 1.674 | -0.0178 | No | ||
| 25 | GPR65 | 3826 | 1.640 | -0.0176 | No | ||
| 26 | TNFSF15 | 3834 | 1.637 | -0.0144 | No | ||
| 27 | CHKA | 3859 | 1.623 | -0.0121 | No | ||
| 28 | THBS1 | 4184 | 1.481 | -0.0263 | No | ||
| 29 | GPSN2 | 4211 | 1.467 | -0.0245 | No | ||
| 30 | RANGAP1 | 4252 | 1.449 | -0.0234 | No | ||
| 31 | GATA6 | 4291 | 1.432 | -0.0223 | No | ||
| 32 | KPNA3 | 4550 | 1.334 | -0.0333 | No | ||
| 33 | UNC5C | 4627 | 1.305 | -0.0345 | No | ||
| 34 | PAX2 | 4793 | 1.236 | -0.0407 | No | ||
| 35 | CDK6 | 4816 | 1.226 | -0.0392 | No | ||
| 36 | BDNF | 4945 | 1.182 | -0.0435 | No | ||
| 37 | KCNE4 | 5113 | 1.136 | -0.0500 | No | ||
| 38 | IL27 | 5205 | 1.105 | -0.0525 | No | ||
| 39 | TNFRSF17 | 5287 | 1.086 | -0.0544 | No | ||
| 40 | COL12A1 | 5433 | 1.033 | -0.0600 | No | ||
| 41 | MIA2 | 5698 | 0.954 | -0.0722 | No | ||
| 42 | PCDH7 | 5829 | 0.918 | -0.0772 | No | ||
| 43 | ZCCHC5 | 5988 | 0.871 | -0.0838 | No | ||
| 44 | PROK2 | 6054 | 0.852 | -0.0854 | No | ||
| 45 | MMP25 | 6151 | 0.825 | -0.0888 | No | ||
| 46 | TNFSF13B | 6304 | 0.786 | -0.0953 | No | ||
| 47 | EREG | 6381 | 0.769 | -0.0977 | No | ||
| 48 | WNT8B | 6608 | 0.708 | -0.1083 | No | ||
| 49 | IFIT2 | 6779 | 0.664 | -0.1161 | No | ||
| 50 | IL22RA1 | 6780 | 0.664 | -0.1146 | No | ||
| 51 | TOP1 | 6905 | 0.631 | -0.1199 | No | ||
| 52 | KCNJ16 | 6908 | 0.631 | -0.1186 | No | ||
| 53 | IFNB1 | 6917 | 0.628 | -0.1176 | No | ||
| 54 | TNRC4 | 6971 | 0.613 | -0.1191 | No | ||
| 55 | PITX3 | 6988 | 0.609 | -0.1187 | No | ||
| 56 | LRP2 | 7283 | 0.541 | -0.1334 | No | ||
| 57 | ZFPM1 | 7417 | 0.508 | -0.1395 | No | ||
| 58 | PLXNC1 | 8169 | 0.312 | -0.1794 | No | ||
| 59 | AMMECR1 | 8279 | 0.282 | -0.1847 | No | ||
| 60 | PLAG1 | 8396 | 0.256 | -0.1904 | No | ||
| 61 | OTX1 | 8421 | 0.252 | -0.1912 | No | ||
| 62 | NPR3 | 8542 | 0.221 | -0.1972 | No | ||
| 63 | CXCL11 | 8643 | 0.198 | -0.2021 | No | ||
| 64 | HTR2C | 8690 | 0.187 | -0.2042 | No | ||
| 65 | EYA4 | 8909 | 0.136 | -0.2157 | No | ||
| 66 | NKX2-2 | 8919 | 0.133 | -0.2159 | No | ||
| 67 | CACNA2D2 | 9023 | 0.105 | -0.2213 | No | ||
| 68 | CRY1 | 9475 | -0.015 | -0.2456 | No | ||
| 69 | ERG | 9592 | -0.046 | -0.2518 | No | ||
| 70 | SLC24A1 | 9689 | -0.070 | -0.2569 | No | ||
| 71 | ADAM15 | 9690 | -0.070 | -0.2567 | No | ||
| 72 | SDPR | 9778 | -0.093 | -0.2612 | No | ||
| 73 | PRDM16 | 9881 | -0.121 | -0.2665 | No | ||
| 74 | OSBP | 10052 | -0.166 | -0.2753 | No | ||
| 75 | SLC12A7 | 10094 | -0.176 | -0.2771 | No | ||
| 76 | TCF15 | 10291 | -0.218 | -0.2873 | No | ||
| 77 | TBX21 | 10337 | -0.229 | -0.2892 | No | ||
| 78 | MSX1 | 10389 | -0.237 | -0.2914 | No | ||
| 79 | PURA | 10543 | -0.271 | -0.2991 | No | ||
| 80 | FIGN | 10823 | -0.338 | -0.3135 | No | ||
| 81 | ERBB2 | 11657 | -0.549 | -0.3574 | No | ||
| 82 | CBX4 | 11856 | -0.607 | -0.3667 | No | ||
| 83 | CXXC5 | 11957 | -0.635 | -0.3707 | No | ||
| 84 | ETV6 | 12055 | -0.663 | -0.3745 | No | ||
| 85 | LMO1 | 12324 | -0.742 | -0.3874 | No | ||
| 86 | OPTN | 12674 | -0.850 | -0.4044 | No | ||
| 87 | ASPA | 12687 | -0.854 | -0.4031 | No | ||
| 88 | SIX1 | 12740 | -0.871 | -0.4040 | No | ||
| 89 | SEMA6D | 12904 | -0.917 | -0.4108 | No | ||
| 90 | LGP2 | 13021 | -0.950 | -0.4150 | No | ||
| 91 | MET | 13134 | -0.992 | -0.4188 | No | ||
| 92 | CDH3 | 13174 | -1.008 | -0.4187 | No | ||
| 93 | OVOL1 | 13545 | -1.147 | -0.4362 | No | ||
| 94 | LSP1 | 13566 | -1.156 | -0.4347 | No | ||
| 95 | ZBP1 | 13687 | -1.204 | -0.4385 | No | ||
| 96 | KYNU | 13702 | -1.211 | -0.4366 | No | ||
| 97 | CXCL10 | 13979 | -1.314 | -0.4486 | No | ||
| 98 | JARID2 | 13982 | -1.315 | -0.4458 | No | ||
| 99 | NT5C3 | 14103 | -1.363 | -0.4493 | No | ||
| 100 | BBX | 14500 | -1.521 | -0.4674 | No | ||
| 101 | DGKA | 14683 | -1.621 | -0.4736 | No | ||
| 102 | HPCAL1 | 14887 | -1.735 | -0.4807 | No | ||
| 103 | ZNF385 | 15041 | -1.822 | -0.4850 | No | ||
| 104 | EGFL6 | 15093 | -1.854 | -0.4836 | No | ||
| 105 | PPARGC1A | 15223 | -1.951 | -0.4863 | No | ||
| 106 | B2M | 15376 | -2.073 | -0.4899 | No | ||
| 107 | RIPK2 | 15542 | -2.199 | -0.4939 | Yes | ||
| 108 | ESR1 | 15580 | -2.232 | -0.4910 | Yes | ||
| 109 | GPX1 | 15584 | -2.235 | -0.4862 | Yes | ||
| 110 | PIGR | 15657 | -2.298 | -0.4850 | Yes | ||
| 111 | YWHAG | 15683 | -2.321 | -0.4812 | Yes | ||
| 112 | USP18 | 15989 | -2.653 | -0.4918 | Yes | ||
| 113 | MAP2K6 | 16017 | -2.684 | -0.4873 | Yes | ||
| 114 | UBE1L | 16064 | -2.736 | -0.4837 | Yes | ||
| 115 | BCL11A | 16072 | -2.744 | -0.4780 | Yes | ||
| 116 | TCIRG1 | 16075 | -2.748 | -0.4720 | Yes | ||
| 117 | FGF9 | 16177 | -2.879 | -0.4710 | Yes | ||
| 118 | PRKACA | 16209 | -2.921 | -0.4662 | Yes | ||
| 119 | TAPBP | 16225 | -2.932 | -0.4605 | Yes | ||
| 120 | MYB | 16420 | -3.240 | -0.4638 | Yes | ||
| 121 | SLC11A2 | 16597 | -3.553 | -0.4654 | Yes | ||
| 122 | GPR18 | 16679 | -3.705 | -0.4616 | Yes | ||
| 123 | USF1 | 16724 | -3.777 | -0.4556 | Yes | ||
| 124 | HOMER1 | 16861 | -4.045 | -0.4539 | Yes | ||
| 125 | TCF7L2 | 16898 | -4.128 | -0.4467 | Yes | ||
| 126 | SH3BGRL | 16960 | -4.275 | -0.4405 | Yes | ||
| 127 | MEIS1 | 16979 | -4.327 | -0.4319 | Yes | ||
| 128 | GSDMDC1 | 16985 | -4.338 | -0.4225 | Yes | ||
| 129 | GNB4 | 17014 | -4.423 | -0.4142 | Yes | ||
| 130 | BLNK | 17018 | -4.436 | -0.4045 | Yes | ||
| 131 | PPP2R5C | 17310 | -5.261 | -0.4085 | Yes | ||
| 132 | STAT6 | 17447 | -5.689 | -0.4033 | Yes | ||
| 133 | MAP3K11 | 17604 | -6.248 | -0.3978 | Yes | ||
| 134 | IRF2 | 17652 | -6.380 | -0.3862 | Yes | ||
| 135 | DAPP1 | 17695 | -6.511 | -0.3740 | Yes | ||
| 136 | PML | 17701 | -6.528 | -0.3597 | Yes | ||
| 137 | HIST1H1C | 17767 | -6.778 | -0.3482 | Yes | ||
| 138 | THRAP1 | 17894 | -7.432 | -0.3385 | Yes | ||
| 139 | CDKN2C | 18019 | -8.014 | -0.3274 | Yes | ||
| 140 | DDX58 | 18077 | -8.456 | -0.3117 | Yes | ||
| 141 | ISGF3G | 18156 | -9.078 | -0.2957 | Yes | ||
| 142 | MS4A1 | 18234 | -9.866 | -0.2780 | Yes | ||
| 143 | CXCR4 | 18241 | -9.911 | -0.2562 | Yes | ||
| 144 | SSBP2 | 18255 | -10.037 | -0.2346 | Yes | ||
| 145 | EHD4 | 18306 | -10.611 | -0.2138 | Yes | ||
| 146 | CD151 | 18319 | -10.828 | -0.1903 | Yes | ||
| 147 | MEF2C | 18390 | -12.088 | -0.1672 | Yes | ||
| 148 | SLA | 18451 | -13.415 | -0.1407 | Yes | ||
| 149 | EGR2 | 18453 | -13.500 | -0.1107 | Yes | ||
| 150 | JMJD2A | 18496 | -15.447 | -0.0786 | Yes | ||
| 151 | DUSP10 | 18536 | -19.103 | -0.0383 | Yes | ||
| 152 | ETV5 | 18537 | -19.144 | 0.0043 | Yes |