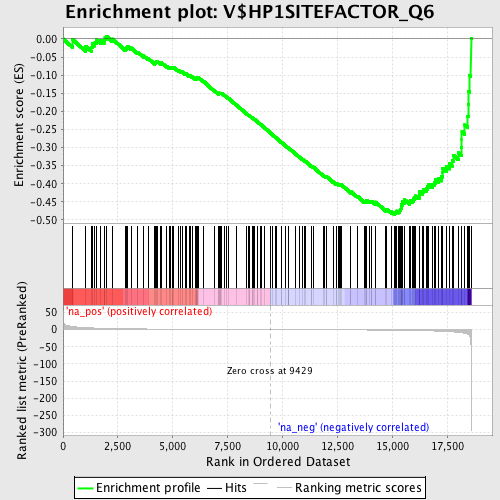
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$HP1SITEFACTOR_Q6 |
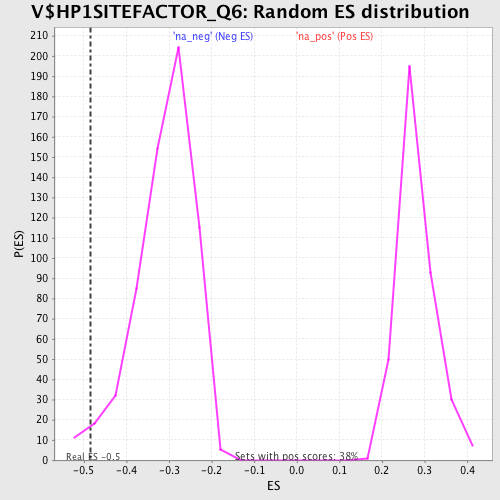
| Enrichment Score (ES) | -0.48491696 |
| Normalized Enrichment Score (NES) | -1.5576996 |
| Nominal p-value | 0.02724359 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SYNCRIP | 429 | 8.944 | -0.0016 | No | ||
| 2 | CDC2L2 | 1035 | 5.684 | -0.0206 | No | ||
| 3 | LUC7L | 1311 | 4.817 | -0.0238 | No | ||
| 4 | KIF1B | 1325 | 4.768 | -0.0129 | No | ||
| 5 | EML4 | 1451 | 4.381 | -0.0091 | No | ||
| 6 | MTCH2 | 1525 | 4.226 | -0.0028 | No | ||
| 7 | MRPL28 | 1714 | 3.831 | -0.0037 | No | ||
| 8 | SFRS2 | 1866 | 3.517 | -0.0033 | No | ||
| 9 | BHLHB3 | 1908 | 3.447 | 0.0028 | No | ||
| 10 | PRICKLE1 | 1968 | 3.356 | 0.0078 | No | ||
| 11 | LMNA | 2246 | 2.920 | -0.0002 | No | ||
| 12 | DMD | 2839 | 2.319 | -0.0266 | No | ||
| 13 | MYH2 | 2906 | 2.256 | -0.0247 | No | ||
| 14 | PHF16 | 2921 | 2.244 | -0.0200 | No | ||
| 15 | NFIX | 3105 | 2.101 | -0.0248 | No | ||
| 16 | PAPD1 | 3404 | 1.874 | -0.0365 | No | ||
| 17 | BARHL1 | 3670 | 1.727 | -0.0466 | No | ||
| 18 | FMO3 | 3870 | 1.616 | -0.0535 | No | ||
| 19 | HOXB3 | 4180 | 1.482 | -0.0666 | No | ||
| 20 | ACRV1 | 4213 | 1.466 | -0.0648 | No | ||
| 21 | LHX6 | 4241 | 1.455 | -0.0627 | No | ||
| 22 | GCNT2 | 4299 | 1.429 | -0.0624 | No | ||
| 23 | NCOR1 | 4438 | 1.378 | -0.0665 | No | ||
| 24 | MYCBP | 4466 | 1.368 | -0.0647 | No | ||
| 25 | BNC2 | 4711 | 1.271 | -0.0748 | No | ||
| 26 | NR5A2 | 4833 | 1.219 | -0.0784 | No | ||
| 27 | STC1 | 4879 | 1.204 | -0.0779 | No | ||
| 28 | HOXD3 | 4963 | 1.176 | -0.0796 | No | ||
| 29 | EVI1 | 4988 | 1.170 | -0.0780 | No | ||
| 30 | CA4 | 5051 | 1.157 | -0.0786 | No | ||
| 31 | GPR156 | 5267 | 1.091 | -0.0876 | No | ||
| 32 | SOX2 | 5352 | 1.065 | -0.0895 | No | ||
| 33 | ZIC1 | 5427 | 1.034 | -0.0910 | No | ||
| 34 | ADAM11 | 5579 | 0.990 | -0.0968 | No | ||
| 35 | VLDLR | 5605 | 0.984 | -0.0958 | No | ||
| 36 | POU4F1 | 5741 | 0.943 | -0.1008 | No | ||
| 37 | POLK | 5788 | 0.928 | -0.1011 | No | ||
| 38 | LHX1 | 5897 | 0.898 | -0.1047 | No | ||
| 39 | PROK2 | 6054 | 0.852 | -0.1111 | No | ||
| 40 | JAG1 | 6066 | 0.848 | -0.1097 | No | ||
| 41 | PITX2 | 6070 | 0.848 | -0.1078 | No | ||
| 42 | PCDHA7 | 6124 | 0.832 | -0.1086 | No | ||
| 43 | CACNA2D3 | 6125 | 0.832 | -0.1066 | No | ||
| 44 | FBN2 | 6166 | 0.822 | -0.1068 | No | ||
| 45 | GDF8 | 6392 | 0.767 | -0.1171 | No | ||
| 46 | CSF3 | 6916 | 0.629 | -0.1439 | No | ||
| 47 | FBXL19 | 7082 | 0.590 | -0.1514 | No | ||
| 48 | SCHIP1 | 7084 | 0.590 | -0.1501 | No | ||
| 49 | FAP | 7115 | 0.580 | -0.1503 | No | ||
| 50 | PJA1 | 7123 | 0.578 | -0.1493 | No | ||
| 51 | USP13 | 7170 | 0.567 | -0.1504 | No | ||
| 52 | HSD17B2 | 7193 | 0.562 | -0.1502 | No | ||
| 53 | RDH10 | 7214 | 0.555 | -0.1499 | No | ||
| 54 | SEZ6 | 7333 | 0.529 | -0.1551 | No | ||
| 55 | IGF1 | 7424 | 0.506 | -0.1587 | No | ||
| 56 | FOXA2 | 7558 | 0.471 | -0.1648 | No | ||
| 57 | ANPEP | 7919 | 0.376 | -0.1834 | No | ||
| 58 | GPR110 | 8350 | 0.267 | -0.2060 | No | ||
| 59 | AFP | 8441 | 0.249 | -0.2103 | No | ||
| 60 | AXL | 8485 | 0.237 | -0.2120 | No | ||
| 61 | LEMD1 | 8486 | 0.237 | -0.2115 | No | ||
| 62 | LRFN5 | 8619 | 0.204 | -0.2181 | No | ||
| 63 | H2AFY | 8627 | 0.202 | -0.2180 | No | ||
| 64 | SFXN5 | 8685 | 0.187 | -0.2207 | No | ||
| 65 | XPO1 | 8722 | 0.178 | -0.2222 | No | ||
| 66 | NOG | 8841 | 0.153 | -0.2282 | No | ||
| 67 | RBP4 | 8857 | 0.147 | -0.2286 | No | ||
| 68 | GPR27 | 8998 | 0.113 | -0.2360 | No | ||
| 69 | TYRO3 | 9032 | 0.104 | -0.2375 | No | ||
| 70 | RIN2 | 9161 | 0.066 | -0.2443 | No | ||
| 71 | IBRDC2 | 9446 | -0.006 | -0.2597 | No | ||
| 72 | NCALD | 9466 | -0.013 | -0.2606 | No | ||
| 73 | DLX1 | 9542 | -0.034 | -0.2646 | No | ||
| 74 | DEPDC2 | 9668 | -0.064 | -0.2712 | No | ||
| 75 | ACCN4 | 9717 | -0.076 | -0.2737 | No | ||
| 76 | SNCAIP | 9936 | -0.136 | -0.2851 | No | ||
| 77 | HAPLN2 | 9953 | -0.140 | -0.2857 | No | ||
| 78 | CS | 10116 | -0.181 | -0.2940 | No | ||
| 79 | UBXD3 | 10250 | -0.211 | -0.3007 | No | ||
| 80 | TSHB | 10278 | -0.216 | -0.3016 | No | ||
| 81 | RHOQ | 10293 | -0.218 | -0.3019 | No | ||
| 82 | EIF4G2 | 10595 | -0.284 | -0.3175 | No | ||
| 83 | HOXB7 | 10776 | -0.327 | -0.3265 | No | ||
| 84 | FLRT1 | 10923 | -0.367 | -0.3335 | No | ||
| 85 | CRYGD | 10983 | -0.381 | -0.3358 | No | ||
| 86 | RCOR1 | 11062 | -0.399 | -0.3390 | No | ||
| 87 | CHRDL1 | 11318 | -0.466 | -0.3517 | No | ||
| 88 | POU3F3 | 11335 | -0.469 | -0.3514 | No | ||
| 89 | ATP6V1E2 | 11399 | -0.483 | -0.3537 | No | ||
| 90 | PIK3R1 | 11865 | -0.608 | -0.3774 | No | ||
| 91 | HOXA9 | 11930 | -0.627 | -0.3793 | No | ||
| 92 | ATP2B3 | 11984 | -0.644 | -0.3807 | No | ||
| 93 | SPARC | 12010 | -0.650 | -0.3804 | No | ||
| 94 | TBX2 | 12307 | -0.737 | -0.3947 | No | ||
| 95 | NPAS3 | 12464 | -0.785 | -0.4012 | No | ||
| 96 | DIO2 | 12477 | -0.792 | -0.4000 | No | ||
| 97 | PPP1CB | 12555 | -0.817 | -0.4022 | No | ||
| 98 | GPC3 | 12612 | -0.833 | -0.4032 | No | ||
| 99 | DMTF1 | 12621 | -0.836 | -0.4016 | No | ||
| 100 | ESRRG | 12675 | -0.851 | -0.4024 | No | ||
| 101 | HOXA4 | 13107 | -0.980 | -0.4234 | No | ||
| 102 | MAP4K4 | 13118 | -0.983 | -0.4215 | No | ||
| 103 | HOXA11 | 13400 | -1.088 | -0.4341 | No | ||
| 104 | CYFIP2 | 13748 | -1.231 | -0.4499 | No | ||
| 105 | DRD3 | 13769 | -1.238 | -0.4480 | No | ||
| 106 | FOXD3 | 13823 | -1.256 | -0.4478 | No | ||
| 107 | FOXP2 | 13833 | -1.260 | -0.4453 | No | ||
| 108 | NRXN3 | 13957 | -1.305 | -0.4488 | No | ||
| 109 | CNTFR | 14034 | -1.336 | -0.4496 | No | ||
| 110 | CALCRL | 14074 | -1.351 | -0.4485 | No | ||
| 111 | ADAM28 | 14235 | -1.410 | -0.4537 | No | ||
| 112 | STAG2 | 14238 | -1.413 | -0.4504 | No | ||
| 113 | BMP5 | 14704 | -1.629 | -0.4717 | No | ||
| 114 | POU4F2 | 14748 | -1.653 | -0.4700 | No | ||
| 115 | DPYSL2 | 14959 | -1.772 | -0.4771 | No | ||
| 116 | OFCC1 | 15105 | -1.864 | -0.4804 | Yes | ||
| 117 | ELF4 | 15152 | -1.898 | -0.4783 | Yes | ||
| 118 | TNNC1 | 15183 | -1.920 | -0.4753 | Yes | ||
| 119 | H3F3B | 15288 | -2.007 | -0.4760 | Yes | ||
| 120 | MAPK10 | 15352 | -2.059 | -0.4744 | Yes | ||
| 121 | TNKS1BP1 | 15361 | -2.065 | -0.4699 | Yes | ||
| 122 | CHD2 | 15400 | -2.088 | -0.4669 | Yes | ||
| 123 | RGS3 | 15418 | -2.102 | -0.4627 | Yes | ||
| 124 | PUM2 | 15427 | -2.108 | -0.4580 | Yes | ||
| 125 | A2BP1 | 15449 | -2.126 | -0.4540 | Yes | ||
| 126 | PARD6A | 15467 | -2.142 | -0.4497 | Yes | ||
| 127 | POU2F1 | 15541 | -2.198 | -0.4483 | Yes | ||
| 128 | HIVEP3 | 15570 | -2.221 | -0.4445 | Yes | ||
| 129 | AP1S2 | 15797 | -2.432 | -0.4508 | Yes | ||
| 130 | ZBTB10 | 15828 | -2.459 | -0.4465 | Yes | ||
| 131 | TGFB3 | 15935 | -2.580 | -0.4460 | Yes | ||
| 132 | CASP8 | 15978 | -2.634 | -0.4418 | Yes | ||
| 133 | MAP2K6 | 16017 | -2.684 | -0.4374 | Yes | ||
| 134 | BCL11A | 16072 | -2.744 | -0.4337 | Yes | ||
| 135 | TAPBP | 16225 | -2.932 | -0.4348 | Yes | ||
| 136 | COL4A3BP | 16242 | -2.954 | -0.4285 | Yes | ||
| 137 | PLS3 | 16252 | -2.966 | -0.4218 | Yes | ||
| 138 | AP1G1 | 16381 | -3.175 | -0.4210 | Yes | ||
| 139 | ZADH2 | 16436 | -3.272 | -0.4160 | Yes | ||
| 140 | CDKL5 | 16550 | -3.473 | -0.4137 | Yes | ||
| 141 | CTCF | 16596 | -3.552 | -0.4075 | Yes | ||
| 142 | VAMP3 | 16638 | -3.630 | -0.4010 | Yes | ||
| 143 | RAB3IP | 16828 | -3.975 | -0.4016 | Yes | ||
| 144 | PRKACB | 16911 | -4.157 | -0.3959 | Yes | ||
| 145 | MEIS1 | 16979 | -4.327 | -0.3890 | Yes | ||
| 146 | PRKAG1 | 17100 | -4.681 | -0.3842 | Yes | ||
| 147 | P2RY10 | 17253 | -5.107 | -0.3800 | Yes | ||
| 148 | HDAC9 | 17284 | -5.180 | -0.3691 | Yes | ||
| 149 | MBNL1 | 17291 | -5.195 | -0.3568 | Yes | ||
| 150 | NOSIP | 17470 | -5.794 | -0.3524 | Yes | ||
| 151 | SMAD1 | 17599 | -6.237 | -0.3442 | Yes | ||
| 152 | HHEX | 17732 | -6.662 | -0.3352 | Yes | ||
| 153 | CITED2 | 17784 | -6.851 | -0.3214 | Yes | ||
| 154 | MBNL2 | 18014 | -7.993 | -0.3144 | Yes | ||
| 155 | LMO4 | 18154 | -9.070 | -0.2999 | Yes | ||
| 156 | SCML4 | 18176 | -9.231 | -0.2787 | Yes | ||
| 157 | ZFP36L1 | 18179 | -9.268 | -0.2563 | Yes | ||
| 158 | SOX4 | 18311 | -10.671 | -0.2375 | Yes | ||
| 159 | TLE4 | 18445 | -13.127 | -0.2129 | Yes | ||
| 160 | BCL6 | 18470 | -13.961 | -0.1803 | Yes | ||
| 161 | HBP1 | 18488 | -15.154 | -0.1445 | Yes | ||
| 162 | DUSP10 | 18536 | -19.103 | -0.1007 | Yes | ||
| 163 | LRRN3 | 18590 | -43.284 | 0.0014 | Yes |