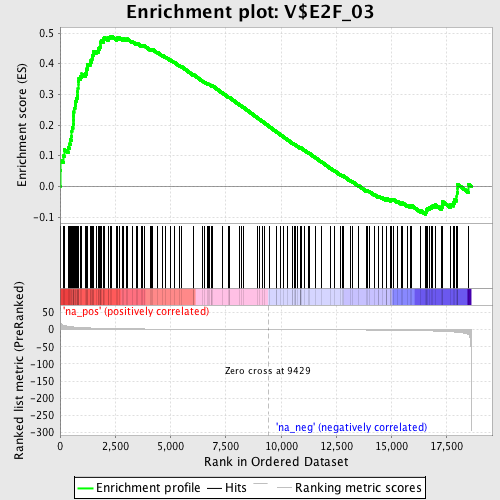
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$E2F_03 |
| Enrichment Score (ES) | 0.48984897 |
| Normalized Enrichment Score (NES) | 1.7935082 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.012900234 |
| FWER p-Value | 0.116 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MCM6 | 7 | 29.907 | 0.0533 | Yes | ||
| 2 | RANBP1 | 36 | 17.971 | 0.0841 | Yes | ||
| 3 | AK2 | 163 | 12.258 | 0.0993 | Yes | ||
| 4 | PPRC1 | 187 | 11.752 | 0.1192 | Yes | ||
| 5 | UNG | 369 | 9.391 | 0.1262 | Yes | ||
| 6 | CDCA7 | 441 | 8.837 | 0.1382 | Yes | ||
| 7 | EIF5A | 471 | 8.526 | 0.1520 | Yes | ||
| 8 | CDC6 | 519 | 8.180 | 0.1641 | Yes | ||
| 9 | POLA2 | 532 | 8.055 | 0.1779 | Yes | ||
| 10 | ORC1L | 539 | 7.998 | 0.1920 | Yes | ||
| 11 | TCP1 | 584 | 7.727 | 0.2035 | Yes | ||
| 12 | LIG1 | 596 | 7.629 | 0.2166 | Yes | ||
| 13 | CDC25A | 605 | 7.562 | 0.2297 | Yes | ||
| 14 | NASP | 618 | 7.483 | 0.2425 | Yes | ||
| 15 | PRKCSH | 629 | 7.405 | 0.2553 | Yes | ||
| 16 | FKBP5 | 675 | 7.146 | 0.2657 | Yes | ||
| 17 | MCM7 | 698 | 7.027 | 0.2771 | Yes | ||
| 18 | MCM3 | 731 | 6.865 | 0.2877 | Yes | ||
| 19 | RAD51 | 776 | 6.691 | 0.2973 | Yes | ||
| 20 | MRPL18 | 800 | 6.617 | 0.3080 | Yes | ||
| 21 | DNMT1 | 808 | 6.587 | 0.3194 | Yes | ||
| 22 | RFC1 | 811 | 6.571 | 0.3311 | Yes | ||
| 23 | ANP32A | 813 | 6.568 | 0.3429 | Yes | ||
| 24 | RPA2 | 849 | 6.392 | 0.3525 | Yes | ||
| 25 | MCM4 | 931 | 6.051 | 0.3589 | Yes | ||
| 26 | MCM2 | 987 | 5.859 | 0.3665 | Yes | ||
| 27 | NUTF2 | 1142 | 5.309 | 0.3677 | Yes | ||
| 28 | GMNN | 1187 | 5.154 | 0.3745 | Yes | ||
| 29 | MAT2A | 1206 | 5.097 | 0.3827 | Yes | ||
| 30 | FUS | 1230 | 5.027 | 0.3905 | Yes | ||
| 31 | KNTC1 | 1246 | 4.968 | 0.3986 | Yes | ||
| 32 | PCSK4 | 1357 | 4.666 | 0.4010 | Yes | ||
| 33 | SMAD6 | 1364 | 4.645 | 0.4091 | Yes | ||
| 34 | CDC45L | 1439 | 4.404 | 0.4130 | Yes | ||
| 35 | SUV39H1 | 1448 | 4.389 | 0.4204 | Yes | ||
| 36 | DNAJC11 | 1467 | 4.353 | 0.4272 | Yes | ||
| 37 | KCNA6 | 1505 | 4.270 | 0.4329 | Yes | ||
| 38 | APRIN | 1509 | 4.258 | 0.4404 | Yes | ||
| 39 | E2F7 | 1632 | 3.991 | 0.4410 | Yes | ||
| 40 | HNRPA1 | 1726 | 3.812 | 0.4428 | Yes | ||
| 41 | PRIM1 | 1730 | 3.806 | 0.4494 | Yes | ||
| 42 | TIAM1 | 1797 | 3.667 | 0.4524 | Yes | ||
| 43 | PRKDC | 1818 | 3.618 | 0.4579 | Yes | ||
| 44 | PRPS1 | 1821 | 3.615 | 0.4642 | Yes | ||
| 45 | E2F3 | 1822 | 3.614 | 0.4707 | Yes | ||
| 46 | SFRS2 | 1866 | 3.517 | 0.4747 | Yes | ||
| 47 | HTF9C | 1952 | 3.379 | 0.4762 | Yes | ||
| 48 | ATAD2 | 1976 | 3.334 | 0.4809 | Yes | ||
| 49 | DLST | 2010 | 3.282 | 0.4850 | Yes | ||
| 50 | SFRS1 | 2175 | 3.032 | 0.4816 | Yes | ||
| 51 | MYC | 2181 | 3.023 | 0.4868 | Yes | ||
| 52 | NR4A2 | 2266 | 2.899 | 0.4874 | Yes | ||
| 53 | CENPB | 2316 | 2.839 | 0.4898 | Yes | ||
| 54 | EIF2S1 | 2562 | 2.558 | 0.4812 | No | ||
| 55 | KCND2 | 2593 | 2.526 | 0.4841 | No | ||
| 56 | DDB2 | 2675 | 2.458 | 0.4841 | No | ||
| 57 | DMD | 2839 | 2.319 | 0.4794 | No | ||
| 58 | NR6A1 | 2881 | 2.279 | 0.4813 | No | ||
| 59 | ZCCHC8 | 2988 | 2.189 | 0.4795 | No | ||
| 60 | E2F1 | 3036 | 2.144 | 0.4808 | No | ||
| 61 | FBXO5 | 3280 | 1.967 | 0.4712 | No | ||
| 62 | SALL1 | 3440 | 1.852 | 0.4659 | No | ||
| 63 | MELK | 3512 | 1.817 | 0.4653 | No | ||
| 64 | AP4M1 | 3666 | 1.730 | 0.4601 | No | ||
| 65 | BRMS1L | 3727 | 1.692 | 0.4599 | No | ||
| 66 | DAXX | 3797 | 1.654 | 0.4591 | No | ||
| 67 | ASXL2 | 4086 | 1.520 | 0.4462 | No | ||
| 68 | PLK4 | 4155 | 1.495 | 0.4452 | No | ||
| 69 | ARID4A | 4176 | 1.483 | 0.4468 | No | ||
| 70 | NR3C2 | 4403 | 1.391 | 0.4371 | No | ||
| 71 | STMN1 | 4636 | 1.302 | 0.4268 | No | ||
| 72 | KCTD15 | 4781 | 1.241 | 0.4213 | No | ||
| 73 | EVI1 | 4988 | 1.170 | 0.4122 | No | ||
| 74 | GLRA3 | 5168 | 1.119 | 0.4045 | No | ||
| 75 | SLBP | 5419 | 1.037 | 0.3928 | No | ||
| 76 | PHF15 | 5494 | 1.017 | 0.3906 | No | ||
| 77 | IPO7 | 6015 | 0.863 | 0.3640 | No | ||
| 78 | KLHDC3 | 6020 | 0.862 | 0.3653 | No | ||
| 79 | CACNA1G | 6462 | 0.746 | 0.3428 | No | ||
| 80 | LGALS1 | 6535 | 0.727 | 0.3402 | No | ||
| 81 | PAQR4 | 6684 | 0.688 | 0.3334 | No | ||
| 82 | ZIC3 | 6699 | 0.686 | 0.3339 | No | ||
| 83 | PTMA | 6756 | 0.671 | 0.3320 | No | ||
| 84 | REPS2 | 6860 | 0.645 | 0.3276 | No | ||
| 85 | GRIA4 | 6872 | 0.641 | 0.3282 | No | ||
| 86 | TOP1 | 6905 | 0.631 | 0.3276 | No | ||
| 87 | SEZ6 | 7333 | 0.529 | 0.3054 | No | ||
| 88 | RPL28 | 7637 | 0.448 | 0.2897 | No | ||
| 89 | PCSK1 | 7664 | 0.441 | 0.2891 | No | ||
| 90 | FANCG | 7678 | 0.437 | 0.2892 | No | ||
| 91 | UXT | 8126 | 0.327 | 0.2656 | No | ||
| 92 | USP52 | 8213 | 0.299 | 0.2614 | No | ||
| 93 | PODN | 8307 | 0.277 | 0.2569 | No | ||
| 94 | SF4 | 8950 | 0.125 | 0.2223 | No | ||
| 95 | DMC1 | 9027 | 0.105 | 0.2184 | No | ||
| 96 | DNAJC9 | 9165 | 0.065 | 0.2111 | No | ||
| 97 | HNRPD | 9228 | 0.051 | 0.2078 | No | ||
| 98 | FMO4 | 9469 | -0.014 | 0.1948 | No | ||
| 99 | PDGFRA | 9795 | -0.098 | 0.1774 | No | ||
| 100 | NUP153 | 9802 | -0.101 | 0.1772 | No | ||
| 101 | TOPBP1 | 9989 | -0.149 | 0.1674 | No | ||
| 102 | SLITRK3 | 10117 | -0.181 | 0.1609 | No | ||
| 103 | TREX2 | 10290 | -0.218 | 0.1519 | No | ||
| 104 | DLL4 | 10514 | -0.263 | 0.1403 | No | ||
| 105 | LUC7L2 | 10521 | -0.265 | 0.1405 | No | ||
| 106 | EIF4G2 | 10595 | -0.284 | 0.1370 | No | ||
| 107 | DCLRE1A | 10637 | -0.292 | 0.1353 | No | ||
| 108 | NRIP3 | 10659 | -0.297 | 0.1347 | No | ||
| 109 | RET | 10762 | -0.322 | 0.1298 | No | ||
| 110 | KLF5 | 10863 | -0.348 | 0.1250 | No | ||
| 111 | RBL1 | 10865 | -0.349 | 0.1256 | No | ||
| 112 | RBBP4 | 10886 | -0.354 | 0.1251 | No | ||
| 113 | EPHB1 | 10895 | -0.356 | 0.1253 | No | ||
| 114 | PLAGL1 | 10907 | -0.360 | 0.1254 | No | ||
| 115 | ARHGEF17 | 11073 | -0.401 | 0.1171 | No | ||
| 116 | HIST1H1D | 11237 | -0.444 | 0.1091 | No | ||
| 117 | HMGN2 | 11287 | -0.455 | 0.1073 | No | ||
| 118 | CCNT2 | 11544 | -0.518 | 0.0943 | No | ||
| 119 | EGLN2 | 11834 | -0.601 | 0.0797 | No | ||
| 120 | RHD | 12231 | -0.710 | 0.0596 | No | ||
| 121 | ATP6V1D | 12406 | -0.768 | 0.0515 | No | ||
| 122 | CASP8AP2 | 12688 | -0.854 | 0.0378 | No | ||
| 123 | RPS20 | 12796 | -0.888 | 0.0336 | No | ||
| 124 | SEMA5A | 12805 | -0.890 | 0.0348 | No | ||
| 125 | BMP7 | 13120 | -0.985 | 0.0195 | No | ||
| 126 | ADAMTS2 | 13246 | -1.032 | 0.0146 | No | ||
| 127 | HNRPA0 | 13525 | -1.139 | 0.0016 | No | ||
| 128 | SLC16A2 | 13874 | -1.274 | -0.0150 | No | ||
| 129 | STAG1 | 13906 | -1.284 | -0.0144 | No | ||
| 130 | TBX3 | 13984 | -1.316 | -0.0162 | No | ||
| 131 | STAG2 | 14238 | -1.413 | -0.0274 | No | ||
| 132 | JPH1 | 14387 | -1.473 | -0.0327 | No | ||
| 133 | H2AFZ | 14415 | -1.488 | -0.0315 | No | ||
| 134 | DIO3 | 14573 | -1.554 | -0.0372 | No | ||
| 135 | ARF3 | 14752 | -1.654 | -0.0439 | No | ||
| 136 | MAZ | 14757 | -1.657 | -0.0412 | No | ||
| 137 | ARHGAP6 | 14779 | -1.669 | -0.0393 | No | ||
| 138 | HRBL | 14946 | -1.763 | -0.0451 | No | ||
| 139 | DPYSL2 | 14959 | -1.772 | -0.0426 | No | ||
| 140 | ANP32E | 15018 | -1.809 | -0.0425 | No | ||
| 141 | TLE3 | 15101 | -1.862 | -0.0436 | No | ||
| 142 | CBX5 | 15290 | -2.008 | -0.0502 | No | ||
| 143 | A2BP1 | 15449 | -2.126 | -0.0549 | No | ||
| 144 | PEG3 | 15502 | -2.164 | -0.0538 | No | ||
| 145 | NHLRC2 | 15740 | -2.371 | -0.0624 | No | ||
| 146 | UFD1L | 15842 | -2.473 | -0.0635 | No | ||
| 147 | AXUD1 | 15918 | -2.559 | -0.0629 | No | ||
| 148 | ATE1 | 16308 | -3.049 | -0.0785 | No | ||
| 149 | CTDSP1 | 16551 | -3.473 | -0.0854 | No | ||
| 150 | HIST1H2AH | 16572 | -3.510 | -0.0802 | No | ||
| 151 | CTCF | 16596 | -3.552 | -0.0751 | No | ||
| 152 | VAMP3 | 16638 | -3.630 | -0.0708 | No | ||
| 153 | PVRL1 | 16732 | -3.784 | -0.0690 | No | ||
| 154 | POLD1 | 16812 | -3.942 | -0.0662 | No | ||
| 155 | RNF121 | 16875 | -4.076 | -0.0622 | No | ||
| 156 | PIM1 | 16981 | -4.328 | -0.0602 | No | ||
| 157 | ING3 | 17279 | -5.165 | -0.0670 | No | ||
| 158 | EZH2 | 17294 | -5.203 | -0.0584 | No | ||
| 159 | FBXL20 | 17308 | -5.257 | -0.0497 | No | ||
| 160 | PPP1R9B | 17682 | -6.443 | -0.0583 | No | ||
| 161 | SLC25A11 | 17790 | -6.885 | -0.0517 | No | ||
| 162 | RAB11B | 17858 | -7.211 | -0.0424 | No | ||
| 163 | RPS6KA5 | 17960 | -7.702 | -0.0341 | No | ||
| 164 | PPM1D | 17963 | -7.713 | -0.0203 | No | ||
| 165 | DCK | 17991 | -7.855 | -0.0077 | No | ||
| 166 | CDKN1A | 17995 | -7.882 | 0.0063 | No | ||
| 167 | MXD3 | 18490 | -15.183 | 0.0068 | No |