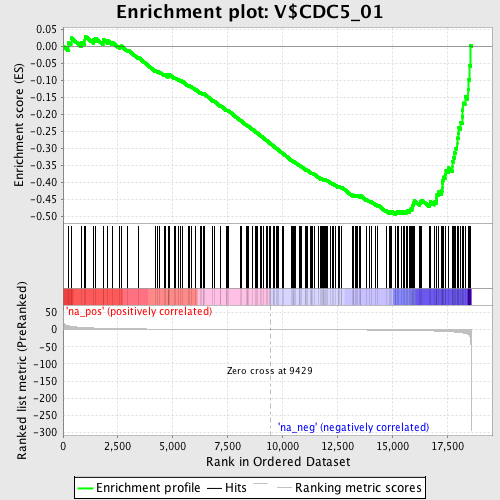
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$CDC5_01 |
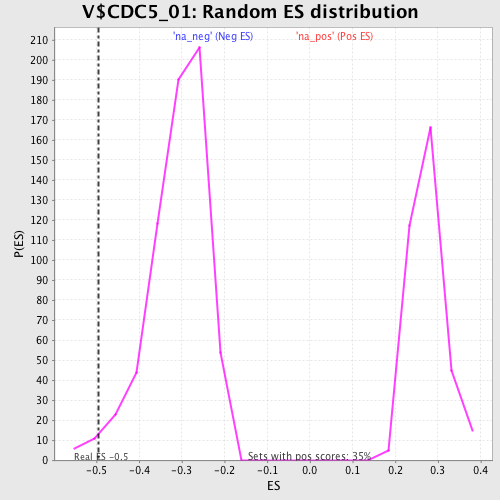
| Enrichment Score (ES) | -0.49548602 |
| Normalized Enrichment Score (NES) | -1.5987598 |
| Nominal p-value | 0.016871165 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SUPT16H | 251 | 10.681 | 0.0098 | No | ||
| 2 | ELP3 | 360 | 9.459 | 0.0246 | No | ||
| 3 | RPA2 | 849 | 6.392 | 0.0121 | No | ||
| 4 | HESX1 | 980 | 5.884 | 0.0179 | No | ||
| 5 | TLE1 | 997 | 5.822 | 0.0298 | No | ||
| 6 | ABLIM1 | 1381 | 4.578 | 0.0190 | No | ||
| 7 | PIK3R3 | 1472 | 4.347 | 0.0237 | No | ||
| 8 | PRPS1 | 1821 | 3.615 | 0.0127 | No | ||
| 9 | HMGA1 | 1826 | 3.607 | 0.0204 | No | ||
| 10 | FNBP1 | 2034 | 3.242 | 0.0162 | No | ||
| 11 | LMNA | 2246 | 2.920 | 0.0112 | No | ||
| 12 | GLI3 | 2559 | 2.562 | -0.0001 | No | ||
| 13 | DUSP14 | 2641 | 2.486 | 0.0009 | No | ||
| 14 | GPR87 | 2956 | 2.210 | -0.0113 | No | ||
| 15 | SALL1 | 3440 | 1.852 | -0.0334 | No | ||
| 16 | ATRX | 4191 | 1.476 | -0.0709 | No | ||
| 17 | PCDH8 | 4320 | 1.419 | -0.0747 | No | ||
| 18 | CHRNA5 | 4400 | 1.391 | -0.0760 | No | ||
| 19 | CLDN1 | 4605 | 1.314 | -0.0841 | No | ||
| 20 | SLC12A8 | 4663 | 1.287 | -0.0844 | No | ||
| 21 | KCTD15 | 4781 | 1.241 | -0.0880 | No | ||
| 22 | PAX2 | 4793 | 1.236 | -0.0859 | No | ||
| 23 | PLCB3 | 4807 | 1.229 | -0.0840 | No | ||
| 24 | LIN7A | 4827 | 1.221 | -0.0823 | No | ||
| 25 | HOXD9 | 5079 | 1.148 | -0.0934 | No | ||
| 26 | MYH6 | 5130 | 1.131 | -0.0937 | No | ||
| 27 | KLF12 | 5259 | 1.093 | -0.0982 | No | ||
| 28 | MOS | 5345 | 1.067 | -0.1005 | No | ||
| 29 | ATOH1 | 5442 | 1.031 | -0.1034 | No | ||
| 30 | RRH | 5730 | 0.946 | -0.1169 | No | ||
| 31 | CART1 | 5754 | 0.940 | -0.1161 | No | ||
| 32 | BCL11B | 5852 | 0.909 | -0.1194 | No | ||
| 33 | PROK2 | 6054 | 0.852 | -0.1284 | No | ||
| 34 | SLC6A13 | 6241 | 0.804 | -0.1368 | No | ||
| 35 | HOXD10 | 6295 | 0.788 | -0.1379 | No | ||
| 36 | RBMS1 | 6386 | 0.768 | -0.1411 | No | ||
| 37 | PHOX2B | 6390 | 0.767 | -0.1396 | No | ||
| 38 | INPP4B | 6421 | 0.757 | -0.1396 | No | ||
| 39 | TMSB4X | 6830 | 0.653 | -0.1603 | No | ||
| 40 | RARA | 6883 | 0.637 | -0.1617 | No | ||
| 41 | TRPS1 | 7169 | 0.567 | -0.1759 | No | ||
| 42 | USP13 | 7170 | 0.567 | -0.1747 | No | ||
| 43 | DNAJC5B | 7444 | 0.499 | -0.1884 | No | ||
| 44 | OTP | 7465 | 0.494 | -0.1884 | No | ||
| 45 | HOXC12 | 7480 | 0.488 | -0.1881 | No | ||
| 46 | VANGL1 | 7532 | 0.476 | -0.1898 | No | ||
| 47 | POT1 | 8078 | 0.338 | -0.2186 | No | ||
| 48 | MYL1 | 8113 | 0.330 | -0.2198 | No | ||
| 49 | TCF21 | 8359 | 0.265 | -0.2325 | No | ||
| 50 | PLAG1 | 8396 | 0.256 | -0.2339 | No | ||
| 51 | TENC1 | 8422 | 0.252 | -0.2347 | No | ||
| 52 | PPIL5 | 8468 | 0.241 | -0.2366 | No | ||
| 53 | LRFN5 | 8619 | 0.204 | -0.2443 | No | ||
| 54 | RORA | 8776 | 0.167 | -0.2524 | No | ||
| 55 | NAP1L5 | 8829 | 0.155 | -0.2548 | No | ||
| 56 | ZFPM2 | 8855 | 0.148 | -0.2559 | No | ||
| 57 | EMILIN1 | 8991 | 0.115 | -0.2629 | No | ||
| 58 | MYL2 | 9026 | 0.105 | -0.2646 | No | ||
| 59 | POU3F1 | 9135 | 0.075 | -0.2703 | No | ||
| 60 | BAI3 | 9280 | 0.037 | -0.2780 | No | ||
| 61 | CLASP1 | 9312 | 0.029 | -0.2796 | No | ||
| 62 | GJA3 | 9407 | 0.006 | -0.2847 | No | ||
| 63 | KLHL4 | 9435 | -0.002 | -0.2861 | No | ||
| 64 | OSR2 | 9439 | -0.004 | -0.2863 | No | ||
| 65 | ODZ1 | 9595 | -0.046 | -0.2946 | No | ||
| 66 | NRAS | 9596 | -0.046 | -0.2945 | No | ||
| 67 | ENPP2 | 9611 | -0.050 | -0.2952 | No | ||
| 68 | SNTG1 | 9714 | -0.075 | -0.3005 | No | ||
| 69 | TNFSF10 | 9760 | -0.087 | -0.3028 | No | ||
| 70 | LYN | 9815 | -0.105 | -0.3055 | No | ||
| 71 | RGS1 | 10010 | -0.156 | -0.3157 | No | ||
| 72 | CALD1 | 10058 | -0.167 | -0.3178 | No | ||
| 73 | MSX1 | 10389 | -0.237 | -0.3352 | No | ||
| 74 | TNNI1 | 10473 | -0.254 | -0.3392 | No | ||
| 75 | DLL4 | 10514 | -0.263 | -0.3408 | No | ||
| 76 | IL20 | 10517 | -0.263 | -0.3403 | No | ||
| 77 | PURA | 10543 | -0.271 | -0.3411 | No | ||
| 78 | NDRG4 | 10613 | -0.287 | -0.3442 | No | ||
| 79 | KLF8 | 10765 | -0.324 | -0.3517 | No | ||
| 80 | HOXB7 | 10776 | -0.327 | -0.3515 | No | ||
| 81 | RORB | 10834 | -0.341 | -0.3538 | No | ||
| 82 | FGF12 | 10842 | -0.343 | -0.3535 | No | ||
| 83 | TNNI3K | 11057 | -0.398 | -0.3642 | No | ||
| 84 | RCOR1 | 11062 | -0.399 | -0.3635 | No | ||
| 85 | MUSK | 11101 | -0.408 | -0.3647 | No | ||
| 86 | MGLL | 11143 | -0.420 | -0.3660 | No | ||
| 87 | NKX2-3 | 11279 | -0.454 | -0.3723 | No | ||
| 88 | CHRDL1 | 11318 | -0.466 | -0.3734 | No | ||
| 89 | GPR85 | 11340 | -0.470 | -0.3735 | No | ||
| 90 | CDH6 | 11374 | -0.479 | -0.3742 | No | ||
| 91 | UCHL1 | 11442 | -0.493 | -0.3768 | No | ||
| 92 | BMPR2 | 11649 | -0.548 | -0.3868 | No | ||
| 93 | DNASE1L2 | 11746 | -0.575 | -0.3907 | No | ||
| 94 | SCN7A | 11764 | -0.580 | -0.3904 | No | ||
| 95 | BAMBI | 11809 | -0.594 | -0.3915 | No | ||
| 96 | HIPK1 | 11861 | -0.608 | -0.3929 | No | ||
| 97 | PIK3R1 | 11865 | -0.608 | -0.3917 | No | ||
| 98 | HOXA9 | 11930 | -0.627 | -0.3938 | No | ||
| 99 | HOXD12 | 11943 | -0.631 | -0.3931 | No | ||
| 100 | FGF10 | 11991 | -0.645 | -0.3942 | No | ||
| 101 | PBX1 | 12036 | -0.657 | -0.3952 | No | ||
| 102 | NEK6 | 12177 | -0.697 | -0.4012 | No | ||
| 103 | SYT4 | 12275 | -0.725 | -0.4049 | No | ||
| 104 | DGKB | 12318 | -0.741 | -0.4056 | No | ||
| 105 | KCNK12 | 12417 | -0.771 | -0.4092 | No | ||
| 106 | ADAMTS9 | 12538 | -0.811 | -0.4139 | No | ||
| 107 | BTBD3 | 12567 | -0.821 | -0.4137 | No | ||
| 108 | GPC3 | 12612 | -0.833 | -0.4142 | No | ||
| 109 | ESRRG | 12675 | -0.851 | -0.4157 | No | ||
| 110 | KLF4 | 12693 | -0.858 | -0.4148 | No | ||
| 111 | IL21 | 13181 | -1.010 | -0.4390 | No | ||
| 112 | PDGFA | 13237 | -1.028 | -0.4397 | No | ||
| 113 | FOXA1 | 13249 | -1.033 | -0.4381 | No | ||
| 114 | CREB5 | 13324 | -1.060 | -0.4398 | No | ||
| 115 | NOL4 | 13384 | -1.081 | -0.4406 | No | ||
| 116 | HOXA11 | 13400 | -1.088 | -0.4390 | No | ||
| 117 | HNRPA0 | 13525 | -1.139 | -0.4433 | No | ||
| 118 | CPNE1 | 13560 | -1.153 | -0.4426 | No | ||
| 119 | BACH2 | 13573 | -1.159 | -0.4407 | No | ||
| 120 | FOXP2 | 13833 | -1.260 | -0.4520 | No | ||
| 121 | NRXN3 | 13957 | -1.305 | -0.4558 | No | ||
| 122 | SESN3 | 14061 | -1.345 | -0.4584 | No | ||
| 123 | STAG2 | 14238 | -1.413 | -0.4649 | No | ||
| 124 | BCOR | 14348 | -1.458 | -0.4676 | No | ||
| 125 | NLK | 14717 | -1.635 | -0.4840 | No | ||
| 126 | HOXC6 | 14860 | -1.720 | -0.4879 | No | ||
| 127 | PRDM13 | 14923 | -1.753 | -0.4875 | No | ||
| 128 | PRRX1 | 14979 | -1.791 | -0.4865 | No | ||
| 129 | VDR | 15145 | -1.894 | -0.4913 | Yes | ||
| 130 | COL16A1 | 15171 | -1.912 | -0.4885 | Yes | ||
| 131 | PPARGC1A | 15223 | -1.951 | -0.4870 | Yes | ||
| 132 | H3F3B | 15288 | -2.007 | -0.4861 | Yes | ||
| 133 | CHD2 | 15400 | -2.088 | -0.4875 | Yes | ||
| 134 | NDNL2 | 15507 | -2.166 | -0.4886 | Yes | ||
| 135 | HIVEP3 | 15570 | -2.221 | -0.4871 | Yes | ||
| 136 | NPTX1 | 15649 | -2.290 | -0.4863 | Yes | ||
| 137 | YWHAG | 15683 | -2.321 | -0.4830 | Yes | ||
| 138 | AP1S2 | 15797 | -2.432 | -0.4838 | Yes | ||
| 139 | SLC27A4 | 15813 | -2.450 | -0.4792 | Yes | ||
| 140 | SHH | 15876 | -2.516 | -0.4771 | Yes | ||
| 141 | TGFB3 | 15935 | -2.580 | -0.4746 | Yes | ||
| 142 | HOXC5 | 15939 | -2.582 | -0.4691 | Yes | ||
| 143 | ODF1 | 15957 | -2.601 | -0.4644 | Yes | ||
| 144 | MXI1 | 15985 | -2.644 | -0.4600 | Yes | ||
| 145 | TITF1 | 16006 | -2.673 | -0.4553 | Yes | ||
| 146 | FUT11 | 16262 | -2.983 | -0.4626 | Yes | ||
| 147 | FYN | 16296 | -3.029 | -0.4577 | Yes | ||
| 148 | ITPKB | 16338 | -3.119 | -0.4531 | Yes | ||
| 149 | ANGPT1 | 16704 | -3.740 | -0.4648 | Yes | ||
| 150 | ROD1 | 16733 | -3.785 | -0.4580 | Yes | ||
| 151 | PRKACB | 16911 | -4.157 | -0.4585 | Yes | ||
| 152 | FOXP1 | 17003 | -4.392 | -0.4538 | Yes | ||
| 153 | BIRC4 | 17030 | -4.487 | -0.4454 | Yes | ||
| 154 | CUTL1 | 17037 | -4.513 | -0.4359 | Yes | ||
| 155 | PRKAG1 | 17100 | -4.681 | -0.4290 | Yes | ||
| 156 | PBX2 | 17251 | -5.102 | -0.4260 | Yes | ||
| 157 | ARPC5L | 17272 | -5.157 | -0.4158 | Yes | ||
| 158 | ING3 | 17279 | -5.165 | -0.4048 | Yes | ||
| 159 | MBNL1 | 17291 | -5.195 | -0.3940 | Yes | ||
| 160 | B3GNT6 | 17343 | -5.365 | -0.3850 | Yes | ||
| 161 | FES | 17409 | -5.547 | -0.3764 | Yes | ||
| 162 | ACTR10 | 17449 | -5.697 | -0.3661 | Yes | ||
| 163 | MAML1 | 17549 | -6.072 | -0.3582 | Yes | ||
| 164 | ACSL4 | 17740 | -6.705 | -0.3538 | Yes | ||
| 165 | SEC63 | 17743 | -6.710 | -0.3392 | Yes | ||
| 166 | CITED2 | 17784 | -6.851 | -0.3264 | Yes | ||
| 167 | GFRA1 | 17841 | -7.114 | -0.3139 | Yes | ||
| 168 | EYA1 | 17879 | -7.339 | -0.2998 | Yes | ||
| 169 | STAT3 | 17952 | -7.686 | -0.2869 | Yes | ||
| 170 | SDC1 | 17954 | -7.690 | -0.2702 | Yes | ||
| 171 | ARNTL | 18007 | -7.954 | -0.2556 | Yes | ||
| 172 | CDKN2C | 18019 | -8.014 | -0.2386 | Yes | ||
| 173 | ARRDC3 | 18128 | -8.916 | -0.2250 | Yes | ||
| 174 | CSNK1G3 | 18193 | -9.429 | -0.2078 | Yes | ||
| 175 | HSPA2 | 18208 | -9.540 | -0.1877 | Yes | ||
| 176 | CDC42EP3 | 18225 | -9.787 | -0.1672 | Yes | ||
| 177 | SAMSN1 | 18320 | -10.839 | -0.1485 | Yes | ||
| 178 | EGR2 | 18453 | -13.500 | -0.1262 | Yes | ||
| 179 | BCL6 | 18470 | -13.961 | -0.0965 | Yes | ||
| 180 | TMPRSS3 | 18540 | -19.616 | -0.0573 | Yes | ||
| 181 | AICDA | 18571 | -28.054 | 0.0024 | Yes |