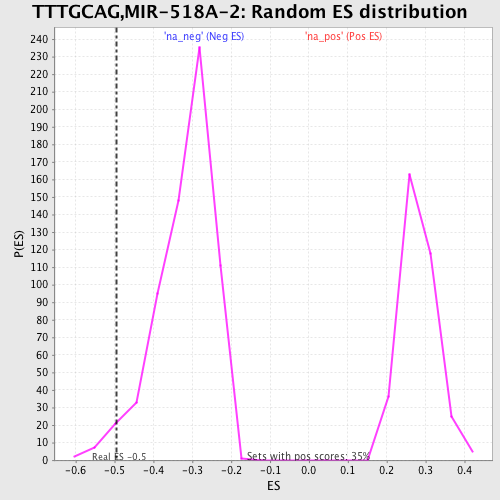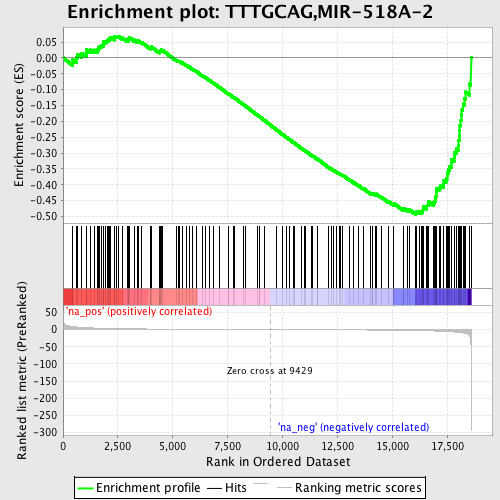
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TTTGCAG,MIR-518A-2 |
| Enrichment Score (ES) | -0.49387744 |
| Normalized Enrichment Score (NES) | -1.5500573 |
| Nominal p-value | 0.030627871 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ETF1 | 442 | 8.832 | -0.0061 | No | ||
| 2 | PICALM | 593 | 7.642 | 0.0012 | No | ||
| 3 | SLC25A26 | 660 | 7.235 | 0.0123 | No | ||
| 4 | RBBP7 | 853 | 6.365 | 0.0147 | No | ||
| 5 | UBAP2 | 1064 | 5.568 | 0.0146 | No | ||
| 6 | PPM1F | 1071 | 5.526 | 0.0255 | No | ||
| 7 | IVNS1ABP | 1236 | 5.009 | 0.0267 | No | ||
| 8 | PABPN1 | 1434 | 4.430 | 0.0250 | No | ||
| 9 | NEDD8 | 1567 | 4.135 | 0.0262 | No | ||
| 10 | BCL2 | 1621 | 4.010 | 0.0314 | No | ||
| 11 | CDK2AP1 | 1666 | 3.924 | 0.0370 | No | ||
| 12 | EIF3S1 | 1759 | 3.752 | 0.0396 | No | ||
| 13 | NCKIPSD | 1823 | 3.612 | 0.0434 | No | ||
| 14 | HMGA1 | 1826 | 3.607 | 0.0506 | No | ||
| 15 | R3HDM1 | 1927 | 3.414 | 0.0521 | No | ||
| 16 | HMGB1 | 2001 | 3.300 | 0.0548 | No | ||
| 17 | ANKRD17 | 2053 | 3.218 | 0.0586 | No | ||
| 18 | SEH1L | 2104 | 3.134 | 0.0622 | No | ||
| 19 | FUSIP1 | 2172 | 3.035 | 0.0647 | No | ||
| 20 | EGR3 | 2326 | 2.825 | 0.0621 | No | ||
| 21 | AEBP2 | 2329 | 2.818 | 0.0677 | No | ||
| 22 | TARDBP | 2412 | 2.704 | 0.0687 | No | ||
| 23 | CORO1C | 2511 | 2.605 | 0.0687 | No | ||
| 24 | CTDP1 | 2709 | 2.430 | 0.0629 | No | ||
| 25 | PRKAB2 | 2920 | 2.244 | 0.0561 | No | ||
| 26 | RHOT1 | 2982 | 2.194 | 0.0572 | No | ||
| 27 | DHX15 | 2987 | 2.190 | 0.0614 | No | ||
| 28 | TOMM70A | 3006 | 2.170 | 0.0648 | No | ||
| 29 | MMD | 3233 | 2.002 | 0.0566 | No | ||
| 30 | TAF4 | 3376 | 1.891 | 0.0528 | No | ||
| 31 | PAX3 | 3429 | 1.858 | 0.0537 | No | ||
| 32 | PNN | 3589 | 1.766 | 0.0487 | No | ||
| 33 | SPG20 | 3973 | 1.570 | 0.0311 | No | ||
| 34 | CATSPER2 | 3988 | 1.563 | 0.0335 | No | ||
| 35 | AFF2 | 4026 | 1.546 | 0.0346 | No | ||
| 36 | ACTR3 | 4382 | 1.398 | 0.0182 | No | ||
| 37 | NR3C2 | 4403 | 1.391 | 0.0200 | No | ||
| 38 | DSCAML1 | 4419 | 1.385 | 0.0219 | No | ||
| 39 | NCOR1 | 4438 | 1.378 | 0.0237 | No | ||
| 40 | LAMC1 | 4463 | 1.371 | 0.0252 | No | ||
| 41 | KPNA3 | 4550 | 1.334 | 0.0233 | No | ||
| 42 | GDF11 | 5154 | 1.122 | -0.0071 | No | ||
| 43 | HMGA2 | 5262 | 1.092 | -0.0107 | No | ||
| 44 | CNN1 | 5289 | 1.086 | -0.0099 | No | ||
| 45 | ZIC1 | 5427 | 1.034 | -0.0153 | No | ||
| 46 | SET | 5617 | 0.980 | -0.0235 | No | ||
| 47 | POU4F1 | 5741 | 0.943 | -0.0283 | No | ||
| 48 | OSBPL8 | 5917 | 0.893 | -0.0359 | No | ||
| 49 | JAG1 | 6066 | 0.848 | -0.0422 | No | ||
| 50 | SGCZ | 6355 | 0.775 | -0.0563 | No | ||
| 51 | DSC3 | 6488 | 0.741 | -0.0619 | No | ||
| 52 | MYOG | 6672 | 0.692 | -0.0704 | No | ||
| 53 | DKK2 | 6861 | 0.645 | -0.0793 | No | ||
| 54 | CREB3L2 | 7117 | 0.580 | -0.0919 | No | ||
| 55 | KCNJ2 | 7543 | 0.474 | -0.1140 | No | ||
| 56 | WDR26 | 7550 | 0.473 | -0.1133 | No | ||
| 57 | EIF4G3 | 7787 | 0.406 | -0.1253 | No | ||
| 58 | PLCB1 | 7808 | 0.401 | -0.1256 | No | ||
| 59 | ACVR2A | 8226 | 0.296 | -0.1476 | No | ||
| 60 | CPEB2 | 8319 | 0.273 | -0.1520 | No | ||
| 61 | DDX42 | 8867 | 0.145 | -0.1813 | No | ||
| 62 | AMACR | 8935 | 0.129 | -0.1847 | No | ||
| 63 | RASGRP4 | 8966 | 0.120 | -0.1861 | No | ||
| 64 | KPNA4 | 9176 | 0.063 | -0.1972 | No | ||
| 65 | BTBD11 | 9737 | -0.081 | -0.2274 | No | ||
| 66 | PTGFRN | 9985 | -0.148 | -0.2405 | No | ||
| 67 | ANK2 | 10170 | -0.192 | -0.2500 | No | ||
| 68 | PTPRF | 10334 | -0.228 | -0.2584 | No | ||
| 69 | DAPK1 | 10479 | -0.256 | -0.2657 | No | ||
| 70 | TMEPAI | 10534 | -0.268 | -0.2681 | No | ||
| 71 | SP8 | 10858 | -0.347 | -0.2849 | No | ||
| 72 | GUCY2C | 10979 | -0.381 | -0.2906 | No | ||
| 73 | SAMD4A | 11060 | -0.399 | -0.2941 | No | ||
| 74 | ATP11A | 11319 | -0.466 | -0.3071 | No | ||
| 75 | SIPA1L2 | 11349 | -0.472 | -0.3077 | No | ||
| 76 | RAI2 | 11573 | -0.527 | -0.3188 | No | ||
| 77 | CUL2 | 11578 | -0.529 | -0.3179 | No | ||
| 78 | STK38 | 12098 | -0.676 | -0.3446 | No | ||
| 79 | SATB2 | 12217 | -0.707 | -0.3496 | No | ||
| 80 | DYRK1A | 12335 | -0.746 | -0.3544 | No | ||
| 81 | ATRN | 12443 | -0.777 | -0.3586 | No | ||
| 82 | THRAP2 | 12576 | -0.823 | -0.3641 | No | ||
| 83 | SEPT6 | 12651 | -0.845 | -0.3664 | No | ||
| 84 | TIMP2 | 12743 | -0.872 | -0.3696 | No | ||
| 85 | SRCRB4D | 13039 | -0.953 | -0.3836 | No | ||
| 86 | FOXA1 | 13249 | -1.033 | -0.3929 | No | ||
| 87 | PLEKHA3 | 13460 | -1.112 | -0.4020 | No | ||
| 88 | DOCK10 | 13689 | -1.205 | -0.4119 | No | ||
| 89 | QKI | 14028 | -1.333 | -0.4275 | No | ||
| 90 | HNRPR | 14107 | -1.365 | -0.4290 | No | ||
| 91 | LEF1 | 14115 | -1.368 | -0.4266 | No | ||
| 92 | STAG2 | 14238 | -1.413 | -0.4303 | No | ||
| 93 | ADRA2B | 14271 | -1.425 | -0.4292 | No | ||
| 94 | PACSIN1 | 14507 | -1.525 | -0.4388 | No | ||
| 95 | NDRG1 | 14814 | -1.689 | -0.4520 | No | ||
| 96 | CHD9 | 15058 | -1.831 | -0.4614 | No | ||
| 97 | RANBP10 | 15069 | -1.841 | -0.4583 | No | ||
| 98 | EIF2C2 | 15505 | -2.165 | -0.4774 | No | ||
| 99 | CEPT1 | 15523 | -2.181 | -0.4740 | No | ||
| 100 | INHBB | 15712 | -2.351 | -0.4794 | No | ||
| 101 | SRPK2 | 15786 | -2.422 | -0.4784 | No | ||
| 102 | BCL11A | 16072 | -2.744 | -0.4883 | Yes | ||
| 103 | SLC39A9 | 16116 | -2.797 | -0.4850 | Yes | ||
| 104 | ARPP-19 | 16229 | -2.939 | -0.4851 | Yes | ||
| 105 | CNNM2 | 16323 | -3.095 | -0.4839 | Yes | ||
| 106 | CGGBP1 | 16373 | -3.168 | -0.4802 | Yes | ||
| 107 | PHF6 | 16403 | -3.222 | -0.4752 | Yes | ||
| 108 | FBXL5 | 16424 | -3.244 | -0.4698 | Yes | ||
| 109 | TMEM32 | 16554 | -3.480 | -0.4697 | Yes | ||
| 110 | RAB22A | 16585 | -3.535 | -0.4642 | Yes | ||
| 111 | PPP1R12A | 16635 | -3.625 | -0.4595 | Yes | ||
| 112 | MAPRE1 | 16662 | -3.675 | -0.4535 | Yes | ||
| 113 | PURB | 16872 | -4.070 | -0.4566 | Yes | ||
| 114 | YTHDF3 | 16942 | -4.235 | -0.4518 | Yes | ||
| 115 | MEIS1 | 16979 | -4.327 | -0.4450 | Yes | ||
| 116 | SURF4 | 16991 | -4.357 | -0.4368 | Yes | ||
| 117 | NRF1 | 17013 | -4.422 | -0.4290 | Yes | ||
| 118 | GNB4 | 17014 | -4.423 | -0.4201 | Yes | ||
| 119 | BIRC4 | 17030 | -4.487 | -0.4118 | Yes | ||
| 120 | PFN2 | 17136 | -4.765 | -0.4079 | Yes | ||
| 121 | CD47 | 17217 | -4.993 | -0.4021 | Yes | ||
| 122 | LAMP2 | 17336 | -5.349 | -0.3977 | Yes | ||
| 123 | MAP3K3 | 17358 | -5.402 | -0.3879 | Yes | ||
| 124 | MKRN1 | 17453 | -5.719 | -0.3815 | Yes | ||
| 125 | RAB6A | 17498 | -5.898 | -0.3719 | Yes | ||
| 126 | RAB21 | 17501 | -5.914 | -0.3601 | Yes | ||
| 127 | KCMF1 | 17569 | -6.123 | -0.3513 | Yes | ||
| 128 | RAP2C | 17622 | -6.298 | -0.3414 | Yes | ||
| 129 | RNF11 | 17690 | -6.488 | -0.3320 | Yes | ||
| 130 | PLEKHB2 | 17694 | -6.501 | -0.3190 | Yes | ||
| 131 | HRB | 17818 | -7.022 | -0.3115 | Yes | ||
| 132 | BRI3 | 17847 | -7.138 | -0.2986 | Yes | ||
| 133 | NEDD4L | 17914 | -7.545 | -0.2869 | Yes | ||
| 134 | PLEKHM1 | 18005 | -7.941 | -0.2757 | Yes | ||
| 135 | PJA2 | 18021 | -8.022 | -0.2603 | Yes | ||
| 136 | ITCH | 18052 | -8.262 | -0.2453 | Yes | ||
| 137 | DUSP6 | 18078 | -8.479 | -0.2295 | Yes | ||
| 138 | WDR22 | 18087 | -8.549 | -0.2127 | Yes | ||
| 139 | ARRDC3 | 18128 | -8.916 | -0.1968 | Yes | ||
| 140 | IHPK1 | 18152 | -9.044 | -0.1798 | Yes | ||
| 141 | ZFP36L1 | 18179 | -9.268 | -0.1625 | Yes | ||
| 142 | CDC42EP3 | 18225 | -9.787 | -0.1451 | Yes | ||
| 143 | GADD45A | 18296 | -10.483 | -0.1277 | Yes | ||
| 144 | TCF12 | 18338 | -11.172 | -0.1074 | Yes | ||
| 145 | JARID1B | 18520 | -17.278 | -0.0823 | Yes | ||
| 146 | LRRN3 | 18590 | -43.284 | 0.0014 | Yes |