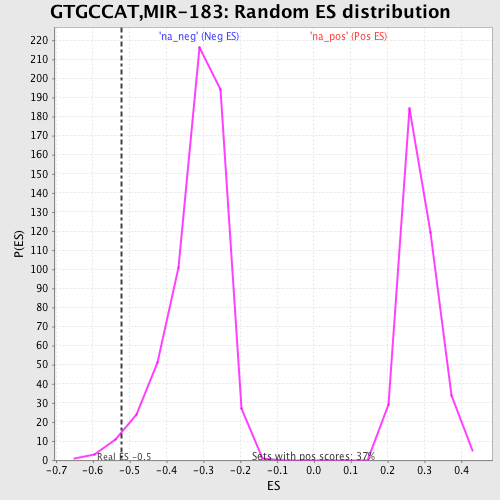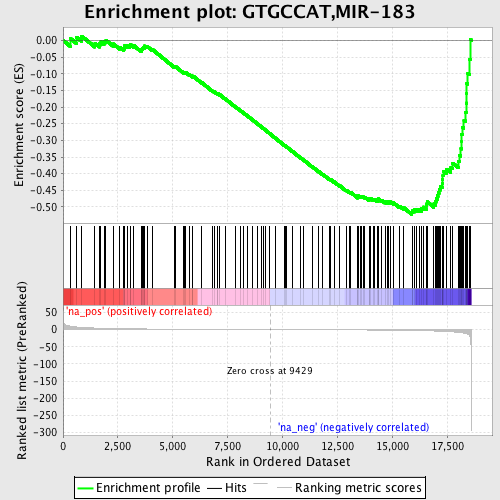
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GTGCCAT,MIR-183 |
| Enrichment Score (ES) | -0.5224945 |
| Normalized Enrichment Score (NES) | -1.637786 |
| Nominal p-value | 0.0190779 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VDAC1 | 348 | 9.561 | 0.0049 | No | ||
| 2 | PPP1R14B | 607 | 7.550 | 0.0097 | No | ||
| 3 | PPP2CB | 844 | 6.412 | 0.0129 | No | ||
| 4 | EML4 | 1451 | 4.381 | -0.0090 | No | ||
| 5 | AMD1 | 1655 | 3.944 | -0.0102 | No | ||
| 6 | PKP4 | 1724 | 3.815 | -0.0044 | No | ||
| 7 | SFRS2 | 1866 | 3.517 | -0.0033 | No | ||
| 8 | LPHN1 | 1951 | 3.380 | 0.0006 | No | ||
| 9 | SOX6 | 2288 | 2.874 | -0.0104 | No | ||
| 10 | L3MBTL3 | 2582 | 2.538 | -0.0200 | No | ||
| 11 | IDH2 | 2774 | 2.372 | -0.0244 | No | ||
| 12 | ITGB1 | 2804 | 2.343 | -0.0202 | No | ||
| 13 | OGT | 2811 | 2.337 | -0.0147 | No | ||
| 14 | FREQ | 2943 | 2.223 | -0.0162 | No | ||
| 15 | PDCD6 | 3048 | 2.137 | -0.0166 | No | ||
| 16 | SIN3A | 3059 | 2.132 | -0.0118 | No | ||
| 17 | DTNA | 3225 | 2.010 | -0.0157 | No | ||
| 18 | PLEKHA5 | 3564 | 1.779 | -0.0296 | No | ||
| 19 | UNC13B | 3590 | 1.766 | -0.0266 | No | ||
| 20 | PLAGL2 | 3609 | 1.755 | -0.0232 | No | ||
| 21 | LRP6 | 3644 | 1.740 | -0.0207 | No | ||
| 22 | RNF138 | 3706 | 1.705 | -0.0197 | No | ||
| 23 | BRMS1L | 3727 | 1.692 | -0.0166 | No | ||
| 24 | TUB | 3824 | 1.641 | -0.0177 | No | ||
| 25 | PLCB4 | 4066 | 1.527 | -0.0270 | No | ||
| 26 | MAL2 | 5068 | 1.151 | -0.0783 | No | ||
| 27 | CSNK1G1 | 5120 | 1.133 | -0.0782 | No | ||
| 28 | PHF15 | 5494 | 1.017 | -0.0959 | No | ||
| 29 | TPM1 | 5534 | 1.004 | -0.0955 | No | ||
| 30 | PAM | 5575 | 0.991 | -0.0952 | No | ||
| 31 | VIL2 | 5769 | 0.935 | -0.1033 | No | ||
| 32 | RAI14 | 5899 | 0.897 | -0.1080 | No | ||
| 33 | OSBPL8 | 5917 | 0.893 | -0.1067 | No | ||
| 34 | GNG5 | 6315 | 0.784 | -0.1263 | No | ||
| 35 | TMSB4X | 6830 | 0.653 | -0.1524 | No | ||
| 36 | ARHGAP21 | 6882 | 0.637 | -0.1536 | No | ||
| 37 | CDK5R1 | 7018 | 0.604 | -0.1594 | No | ||
| 38 | MTMR6 | 7051 | 0.596 | -0.1597 | No | ||
| 39 | BRUNOL6 | 7140 | 0.574 | -0.1630 | No | ||
| 40 | CUGBP2 | 7382 | 0.518 | -0.1747 | No | ||
| 41 | ARHGEF18 | 7867 | 0.389 | -0.2000 | No | ||
| 42 | BZW1 | 8064 | 0.341 | -0.2097 | No | ||
| 43 | ACVR2A | 8226 | 0.296 | -0.2177 | No | ||
| 44 | PLAG1 | 8396 | 0.256 | -0.2262 | No | ||
| 45 | HBEGF | 8652 | 0.195 | -0.2395 | No | ||
| 46 | ZFPM2 | 8855 | 0.148 | -0.2501 | No | ||
| 47 | SMPD3 | 9055 | 0.097 | -0.2606 | No | ||
| 48 | PHLDB2 | 9139 | 0.071 | -0.2649 | No | ||
| 49 | EPHA4 | 9236 | 0.049 | -0.2700 | No | ||
| 50 | GREM2 | 9401 | 0.007 | -0.2788 | No | ||
| 51 | ABCA1 | 9699 | -0.072 | -0.2947 | No | ||
| 52 | ZFYVE26 | 10108 | -0.179 | -0.3163 | No | ||
| 53 | SLITRK3 | 10117 | -0.181 | -0.3163 | No | ||
| 54 | RAB11FIP4 | 10183 | -0.196 | -0.3193 | No | ||
| 55 | MAP3K4 | 10464 | -0.252 | -0.3339 | No | ||
| 56 | SNX1 | 10832 | -0.341 | -0.3529 | No | ||
| 57 | PEX19 | 10974 | -0.379 | -0.3595 | No | ||
| 58 | KCNJ14 | 11345 | -0.471 | -0.3784 | No | ||
| 59 | STK38L | 11655 | -0.549 | -0.3937 | No | ||
| 60 | SLC35A1 | 11826 | -0.599 | -0.4014 | No | ||
| 61 | PTDSS1 | 12145 | -0.688 | -0.4169 | No | ||
| 62 | PTPN4 | 12190 | -0.699 | -0.4176 | No | ||
| 63 | IRS1 | 12354 | -0.755 | -0.4245 | No | ||
| 64 | SNCB | 12606 | -0.832 | -0.4360 | No | ||
| 65 | SEMA6D | 12904 | -0.917 | -0.4498 | No | ||
| 66 | KCNK2 | 13053 | -0.957 | -0.4554 | No | ||
| 67 | TXNDC4 | 13111 | -0.982 | -0.4561 | No | ||
| 68 | CHRD | 13430 | -1.101 | -0.4705 | No | ||
| 69 | CELSR3 | 13431 | -1.102 | -0.4678 | No | ||
| 70 | PLEKHA3 | 13460 | -1.112 | -0.4666 | No | ||
| 71 | BACH2 | 13573 | -1.159 | -0.4697 | No | ||
| 72 | ZDHHC6 | 13591 | -1.166 | -0.4678 | No | ||
| 73 | RCN2 | 13682 | -1.203 | -0.4696 | No | ||
| 74 | RALGDS | 13751 | -1.232 | -0.4702 | No | ||
| 75 | PALM2 | 13946 | -1.299 | -0.4775 | No | ||
| 76 | PDCD4 | 13963 | -1.308 | -0.4751 | No | ||
| 77 | QKI | 14028 | -1.333 | -0.4753 | No | ||
| 78 | HECTD2 | 14146 | -1.378 | -0.4782 | No | ||
| 79 | YES1 | 14205 | -1.402 | -0.4778 | No | ||
| 80 | CLCN3 | 14331 | -1.451 | -0.4810 | No | ||
| 81 | PPP2R2A | 14355 | -1.460 | -0.4786 | No | ||
| 82 | ICK | 14365 | -1.464 | -0.4755 | No | ||
| 83 | AP3M1 | 14530 | -1.538 | -0.4805 | No | ||
| 84 | CTDSPL | 14671 | -1.612 | -0.4841 | No | ||
| 85 | TPM3 | 14777 | -1.669 | -0.4856 | No | ||
| 86 | PDE4D | 14829 | -1.703 | -0.4841 | No | ||
| 87 | LAPTM4A | 14909 | -1.746 | -0.4841 | No | ||
| 88 | TMPO | 15053 | -1.827 | -0.4873 | No | ||
| 89 | ANKRD13C | 15342 | -2.052 | -0.4977 | No | ||
| 90 | EIF2C2 | 15505 | -2.165 | -0.5011 | No | ||
| 91 | DCX | 15901 | -2.540 | -0.5162 | Yes | ||
| 92 | NCK2 | 15934 | -2.580 | -0.5115 | Yes | ||
| 93 | CXXC6 | 16002 | -2.666 | -0.5085 | Yes | ||
| 94 | USP47 | 16090 | -2.772 | -0.5063 | Yes | ||
| 95 | ARPP-19 | 16229 | -2.939 | -0.5065 | Yes | ||
| 96 | CFL2 | 16345 | -3.128 | -0.5049 | Yes | ||
| 97 | PHF6 | 16403 | -3.222 | -0.5000 | Yes | ||
| 98 | CTDSP1 | 16551 | -3.473 | -0.4993 | Yes | ||
| 99 | PPP2CA | 16557 | -3.483 | -0.4910 | Yes | ||
| 100 | ENAH | 16586 | -3.536 | -0.4837 | Yes | ||
| 101 | RAB8B | 16903 | -4.139 | -0.4905 | Yes | ||
| 102 | PIM1 | 16981 | -4.328 | -0.4839 | Yes | ||
| 103 | FOXP1 | 17003 | -4.392 | -0.4741 | Yes | ||
| 104 | NR3C1 | 17064 | -4.576 | -0.4660 | Yes | ||
| 105 | RALA | 17090 | -4.654 | -0.4558 | Yes | ||
| 106 | PFN2 | 17136 | -4.765 | -0.4464 | Yes | ||
| 107 | MXD4 | 17189 | -4.906 | -0.4370 | Yes | ||
| 108 | ING3 | 17279 | -5.165 | -0.4290 | Yes | ||
| 109 | MBNL1 | 17291 | -5.195 | -0.4167 | Yes | ||
| 110 | PPP2R5C | 17310 | -5.261 | -0.4046 | Yes | ||
| 111 | RPS6KA3 | 17324 | -5.301 | -0.3921 | Yes | ||
| 112 | DGCR2 | 17479 | -5.814 | -0.3860 | Yes | ||
| 113 | ZNF289 | 17672 | -6.427 | -0.3804 | Yes | ||
| 114 | PSEN2 | 17754 | -6.742 | -0.3680 | Yes | ||
| 115 | RHOBTB1 | 18022 | -8.032 | -0.3625 | Yes | ||
| 116 | TTC7B | 18067 | -8.348 | -0.3442 | Yes | ||
| 117 | TRAM1 | 18131 | -8.946 | -0.3253 | Yes | ||
| 118 | BNIP3L | 18146 | -9.020 | -0.3037 | Yes | ||
| 119 | PRKCA | 18153 | -9.054 | -0.2815 | Yes | ||
| 120 | CSNK1G3 | 18193 | -9.429 | -0.2602 | Yes | ||
| 121 | FOXO1A | 18269 | -10.133 | -0.2391 | Yes | ||
| 122 | TCF12 | 18338 | -11.172 | -0.2150 | Yes | ||
| 123 | BTG1 | 18371 | -11.636 | -0.1878 | Yes | ||
| 124 | MEF2C | 18390 | -12.088 | -0.1588 | Yes | ||
| 125 | DAP | 18399 | -12.256 | -0.1288 | Yes | ||
| 126 | TLE4 | 18445 | -13.127 | -0.0986 | Yes | ||
| 127 | DUSP10 | 18536 | -19.103 | -0.0560 | Yes | ||
| 128 | EGR1 | 18558 | -24.249 | 0.0031 | Yes |