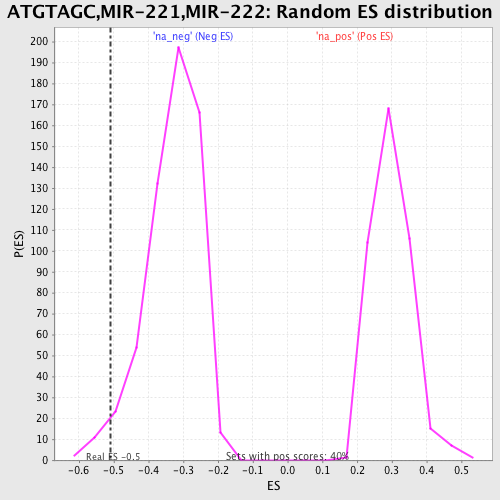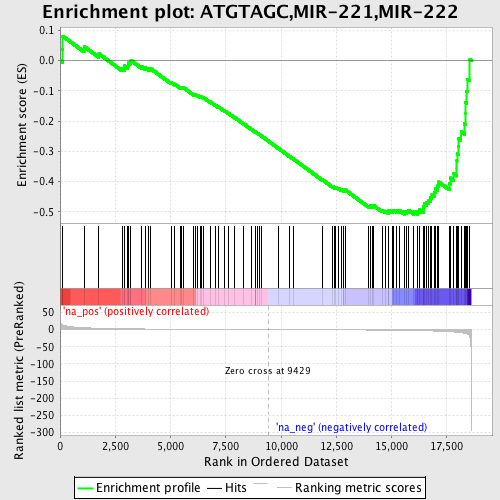
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ATGTAGC,MIR-221,MIR-222 |
| Enrichment Score (ES) | -0.5085447 |
| Normalized Enrichment Score (NES) | -1.5406382 |
| Nominal p-value | 0.028428094 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ACACA | 114 | 13.548 | 0.0377 | No | ||
| 2 | ATP1A1 | 121 | 13.378 | 0.0807 | No | ||
| 3 | CDV3 | 1084 | 5.496 | 0.0466 | No | ||
| 4 | EIF3S1 | 1759 | 3.752 | 0.0223 | No | ||
| 5 | OGT | 2811 | 2.337 | -0.0269 | No | ||
| 6 | HMGCLL1 | 2908 | 2.254 | -0.0247 | No | ||
| 7 | INSIG1 | 2911 | 2.253 | -0.0176 | No | ||
| 8 | ARID1A | 3070 | 2.125 | -0.0192 | No | ||
| 9 | FMR1 | 3089 | 2.112 | -0.0133 | No | ||
| 10 | DACH1 | 3109 | 2.100 | -0.0076 | No | ||
| 11 | POGZ | 3184 | 2.043 | -0.0050 | No | ||
| 12 | NTF3 | 3206 | 2.027 | 0.0005 | No | ||
| 13 | TIMP3 | 3680 | 1.720 | -0.0195 | No | ||
| 14 | PCDHA10 | 3848 | 1.628 | -0.0232 | No | ||
| 15 | AXIN2 | 4018 | 1.547 | -0.0274 | No | ||
| 16 | SLC25A37 | 4095 | 1.517 | -0.0265 | No | ||
| 17 | PLEKHC1 | 5019 | 1.164 | -0.0726 | No | ||
| 18 | MAP3K10 | 5175 | 1.115 | -0.0774 | No | ||
| 19 | PCDHA12 | 5439 | 1.031 | -0.0882 | No | ||
| 20 | IRX5 | 5501 | 1.012 | -0.0882 | No | ||
| 21 | ADAM11 | 5579 | 0.990 | -0.0892 | No | ||
| 22 | PAK1 | 6045 | 0.855 | -0.1115 | No | ||
| 23 | PCDHA7 | 6124 | 0.832 | -0.1131 | No | ||
| 24 | CPNE8 | 6205 | 0.813 | -0.1147 | No | ||
| 25 | UBE2J1 | 6349 | 0.776 | -0.1200 | No | ||
| 26 | CD4 | 6400 | 0.764 | -0.1202 | No | ||
| 27 | KHDRBS2 | 6471 | 0.745 | -0.1215 | No | ||
| 28 | GARNL1 | 6785 | 0.663 | -0.1363 | No | ||
| 29 | EIF5A2 | 7031 | 0.600 | -0.1476 | No | ||
| 30 | TRPS1 | 7169 | 0.567 | -0.1531 | No | ||
| 31 | FNDC3A | 7427 | 0.504 | -0.1654 | No | ||
| 32 | PCDHA3 | 7611 | 0.456 | -0.1738 | No | ||
| 33 | LRFN2 | 7914 | 0.377 | -0.1889 | No | ||
| 34 | AMMECR1 | 8279 | 0.282 | -0.2076 | No | ||
| 35 | PAIP1 | 8663 | 0.192 | -0.2277 | No | ||
| 36 | ARF4 | 8827 | 0.156 | -0.2360 | No | ||
| 37 | NAP1L5 | 8829 | 0.155 | -0.2355 | No | ||
| 38 | ZFPM2 | 8855 | 0.148 | -0.2364 | No | ||
| 39 | SHANK2 | 8938 | 0.129 | -0.2404 | No | ||
| 40 | FOS | 9018 | 0.107 | -0.2443 | No | ||
| 41 | HOXC10 | 9098 | 0.086 | -0.2483 | No | ||
| 42 | NRK | 9887 | -0.124 | -0.2905 | No | ||
| 43 | ATXN1 | 10372 | -0.234 | -0.3158 | No | ||
| 44 | PURA | 10543 | -0.271 | -0.3241 | No | ||
| 45 | HIPK1 | 11861 | -0.608 | -0.3933 | No | ||
| 46 | DYRK1A | 12335 | -0.746 | -0.4164 | No | ||
| 47 | TOX | 12439 | -0.776 | -0.4195 | No | ||
| 48 | PCDHA5 | 12471 | -0.788 | -0.4186 | No | ||
| 49 | SNCB | 12606 | -0.832 | -0.4231 | No | ||
| 50 | RIMS3 | 12746 | -0.873 | -0.4278 | No | ||
| 51 | PCDHA1 | 12822 | -0.893 | -0.4290 | No | ||
| 52 | ETS2 | 12832 | -0.896 | -0.4265 | No | ||
| 53 | SEMA6D | 12904 | -0.917 | -0.4274 | No | ||
| 54 | PCDHA11 | 13971 | -1.311 | -0.4807 | No | ||
| 55 | QKI | 14028 | -1.333 | -0.4794 | No | ||
| 56 | HECTD2 | 14146 | -1.378 | -0.4813 | No | ||
| 57 | MYO10 | 14185 | -1.395 | -0.4788 | No | ||
| 58 | WDR40A | 14579 | -1.556 | -0.4950 | No | ||
| 59 | NLK | 14717 | -1.635 | -0.4971 | No | ||
| 60 | LIFR | 14876 | -1.729 | -0.5000 | Yes | ||
| 61 | RSBN1L | 14879 | -1.731 | -0.4945 | Yes | ||
| 62 | ZNF385 | 15041 | -1.822 | -0.4973 | Yes | ||
| 63 | PCDHA6 | 15091 | -1.853 | -0.4940 | Yes | ||
| 64 | PPARGC1A | 15223 | -1.951 | -0.4947 | Yes | ||
| 65 | MAPK10 | 15352 | -2.059 | -0.4950 | Yes | ||
| 66 | ESR1 | 15580 | -2.232 | -0.5000 | Yes | ||
| 67 | YWHAG | 15683 | -2.321 | -0.4980 | Yes | ||
| 68 | ANGPTL2 | 15755 | -2.387 | -0.4941 | Yes | ||
| 69 | ETS1 | 16011 | -2.677 | -0.4992 | Yes | ||
| 70 | ARNT | 16185 | -2.887 | -0.4992 | Yes | ||
| 71 | VAPB | 16255 | -2.971 | -0.4933 | Yes | ||
| 72 | ZADH2 | 16436 | -3.272 | -0.4924 | Yes | ||
| 73 | PDCD10 | 16453 | -3.298 | -0.4826 | Yes | ||
| 74 | DNAJC14 | 16478 | -3.333 | -0.4731 | Yes | ||
| 75 | CTCF | 16596 | -3.552 | -0.4679 | Yes | ||
| 76 | CBFB | 16690 | -3.725 | -0.4609 | Yes | ||
| 77 | PHF2 | 16772 | -3.867 | -0.4527 | Yes | ||
| 78 | PAIP2 | 16830 | -3.978 | -0.4429 | Yes | ||
| 79 | PPP3R1 | 16929 | -4.205 | -0.4346 | Yes | ||
| 80 | MEIS1 | 16979 | -4.327 | -0.4232 | Yes | ||
| 81 | RALA | 17090 | -4.654 | -0.4141 | Yes | ||
| 82 | DCUN1D1 | 17134 | -4.759 | -0.4010 | Yes | ||
| 83 | CDKN1B | 17623 | -6.299 | -0.4070 | Yes | ||
| 84 | IRF2 | 17652 | -6.380 | -0.3878 | Yes | ||
| 85 | HRB | 17818 | -7.022 | -0.3740 | Yes | ||
| 86 | PPP6C | 17958 | -7.695 | -0.3566 | Yes | ||
| 87 | RAB1A | 17959 | -7.697 | -0.3317 | Yes | ||
| 88 | MESDC1 | 17964 | -7.713 | -0.3069 | Yes | ||
| 89 | CDKN1C | 18027 | -8.074 | -0.2841 | Yes | ||
| 90 | MYLIP | 18045 | -8.182 | -0.2585 | Yes | ||
| 91 | BMF | 18144 | -9.009 | -0.2347 | Yes | ||
| 92 | ASB7 | 18309 | -10.658 | -0.2090 | Yes | ||
| 93 | TCF12 | 18338 | -11.172 | -0.1743 | Yes | ||
| 94 | MIDN | 18342 | -11.212 | -0.1382 | Yes | ||
| 95 | SBK1 | 18401 | -12.359 | -0.1013 | Yes | ||
| 96 | VGLL4 | 18437 | -12.997 | -0.0611 | Yes | ||
| 97 | DMRT3 | 18548 | -21.840 | 0.0037 | Yes |