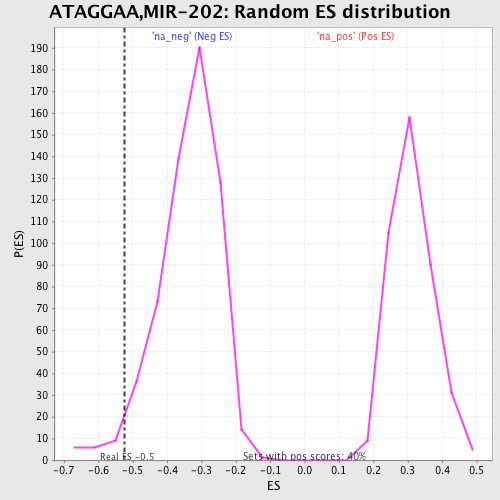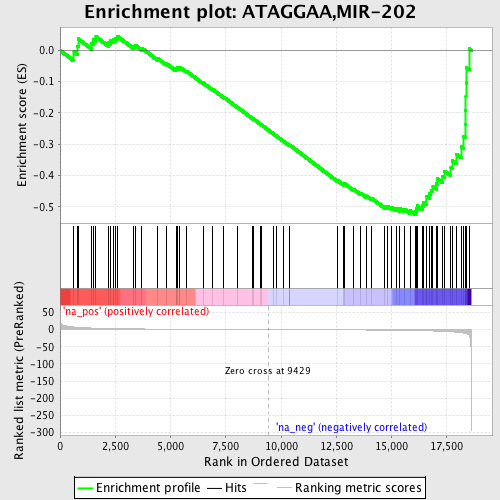
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ATAGGAA,MIR-202 |
| Enrichment Score (ES) | -0.5246293 |
| Normalized Enrichment Score (NES) | -1.5536461 |
| Nominal p-value | 0.03156146 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PPP5C | 627 | 7.420 | -0.0048 | No | ||
| 2 | TTC13 | 772 | 6.706 | 0.0137 | No | ||
| 3 | NARG1 | 821 | 6.535 | 0.0368 | No | ||
| 4 | LUZP1 | 1429 | 4.452 | 0.0215 | No | ||
| 5 | ROBO2 | 1499 | 4.283 | 0.0345 | No | ||
| 6 | BCL2 | 1621 | 4.010 | 0.0437 | No | ||
| 7 | HIP2 | 2204 | 2.995 | 0.0240 | No | ||
| 8 | EAF1 | 2295 | 2.867 | 0.0304 | No | ||
| 9 | TARDBP | 2412 | 2.704 | 0.0347 | No | ||
| 10 | DDX3X | 2518 | 2.596 | 0.0392 | No | ||
| 11 | PPARBP | 2585 | 2.535 | 0.0456 | No | ||
| 12 | DDEF1 | 3327 | 1.926 | 0.0132 | No | ||
| 13 | MIB1 | 3416 | 1.866 | 0.0158 | No | ||
| 14 | SNX16 | 3702 | 1.706 | 0.0071 | No | ||
| 15 | BICD2 | 4409 | 1.389 | -0.0256 | No | ||
| 16 | CTBP2 | 4805 | 1.231 | -0.0420 | No | ||
| 17 | SFPQ | 5245 | 1.097 | -0.0614 | No | ||
| 18 | KLF12 | 5259 | 1.093 | -0.0578 | No | ||
| 19 | IQGAP1 | 5291 | 1.085 | -0.0553 | No | ||
| 20 | USP8 | 5387 | 1.049 | -0.0563 | No | ||
| 21 | CPEB3 | 5412 | 1.040 | -0.0535 | No | ||
| 22 | APPBP2 | 5721 | 0.948 | -0.0664 | No | ||
| 23 | KHDRBS2 | 6471 | 0.745 | -0.1039 | No | ||
| 24 | CD28 | 6895 | 0.633 | -0.1242 | No | ||
| 25 | BAG4 | 7402 | 0.513 | -0.1495 | No | ||
| 26 | KIAA1715 | 8016 | 0.354 | -0.1812 | No | ||
| 27 | DNAJC10 | 8729 | 0.176 | -0.2189 | No | ||
| 28 | PCGF2 | 8743 | 0.174 | -0.2189 | No | ||
| 29 | FAM60A | 9073 | 0.094 | -0.2363 | No | ||
| 30 | HLF | 9105 | 0.085 | -0.2376 | No | ||
| 31 | ACSL3 | 9643 | -0.058 | -0.2663 | No | ||
| 32 | HOXB2 | 9801 | -0.100 | -0.2744 | No | ||
| 33 | RKHD3 | 10125 | -0.183 | -0.2911 | No | ||
| 34 | ATXN1 | 10372 | -0.234 | -0.3035 | No | ||
| 35 | CBL | 10381 | -0.236 | -0.3030 | No | ||
| 36 | MAPK6 | 10388 | -0.237 | -0.3024 | No | ||
| 37 | THRAP2 | 12576 | -0.823 | -0.4171 | No | ||
| 38 | RASA1 | 12828 | -0.895 | -0.4271 | No | ||
| 39 | TRIM33 | 12860 | -0.905 | -0.4252 | No | ||
| 40 | SPRED1 | 13275 | -1.041 | -0.4435 | No | ||
| 41 | SORCS1 | 13617 | -1.180 | -0.4573 | No | ||
| 42 | ARSB | 13862 | -1.271 | -0.4654 | No | ||
| 43 | CNN3 | 14092 | -1.358 | -0.4725 | No | ||
| 44 | ACVR1 | 14685 | -1.621 | -0.4980 | No | ||
| 45 | NDRG1 | 14814 | -1.689 | -0.4983 | No | ||
| 46 | ANP32E | 15018 | -1.809 | -0.5022 | No | ||
| 47 | RAD52 | 15206 | -1.941 | -0.5047 | No | ||
| 48 | EI24 | 15379 | -2.075 | -0.5058 | No | ||
| 49 | ESR1 | 15580 | -2.232 | -0.5079 | No | ||
| 50 | YAF2 | 15837 | -2.468 | -0.5120 | No | ||
| 51 | BCL11A | 16072 | -2.744 | -0.5139 | Yes | ||
| 52 | USP15 | 16132 | -2.823 | -0.5060 | Yes | ||
| 53 | SNAP91 | 16165 | -2.865 | -0.4965 | Yes | ||
| 54 | NR2F2 | 16413 | -3.231 | -0.4972 | Yes | ||
| 55 | GRIA3 | 16434 | -3.270 | -0.4854 | Yes | ||
| 56 | RAB22A | 16585 | -3.535 | -0.4797 | Yes | ||
| 57 | ENAH | 16586 | -3.536 | -0.4658 | Yes | ||
| 58 | ERBB2IP | 16719 | -3.768 | -0.4582 | Yes | ||
| 59 | CREBBP | 16786 | -3.900 | -0.4465 | Yes | ||
| 60 | RNF121 | 16875 | -4.076 | -0.4353 | Yes | ||
| 61 | SENP1 | 17031 | -4.490 | -0.4260 | Yes | ||
| 62 | LBR | 17068 | -4.598 | -0.4100 | Yes | ||
| 63 | RPS6KA3 | 17324 | -5.301 | -0.4030 | Yes | ||
| 64 | PTEN | 17406 | -5.544 | -0.3856 | Yes | ||
| 65 | RNF11 | 17690 | -6.488 | -0.3755 | Yes | ||
| 66 | TGFBR2 | 17742 | -6.708 | -0.3519 | Yes | ||
| 67 | STAT3 | 17952 | -7.686 | -0.3331 | Yes | ||
| 68 | ELF1 | 18149 | -9.026 | -0.3083 | Yes | ||
| 69 | SSBP2 | 18255 | -10.037 | -0.2747 | Yes | ||
| 70 | VANGL2 | 18330 | -11.042 | -0.2354 | Yes | ||
| 71 | TCF12 | 18338 | -11.172 | -0.1920 | Yes | ||
| 72 | ZFAND3 | 18368 | -11.579 | -0.1482 | Yes | ||
| 73 | BTG1 | 18371 | -11.636 | -0.1028 | Yes | ||
| 74 | KRAS | 18407 | -12.449 | -0.0559 | Yes | ||
| 75 | MARCKS | 18519 | -17.134 | 0.0052 | Yes |