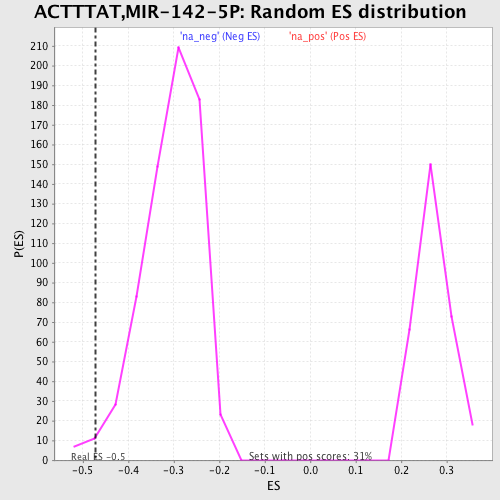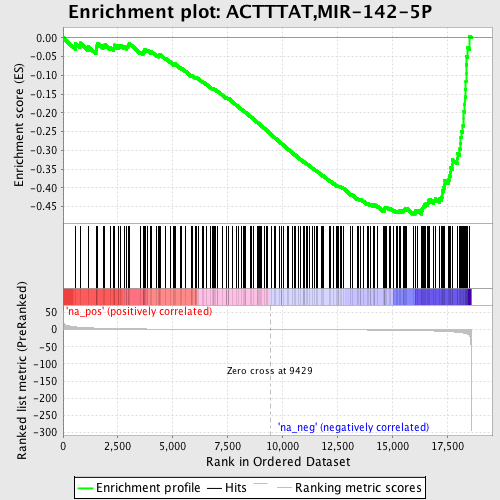
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ACTTTAT,MIR-142-5P |
| Enrichment Score (ES) | -0.47122157 |
| Normalized Enrichment Score (NES) | -1.5439528 |
| Nominal p-value | 0.017316017 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HDLBP | 557 | 7.887 | -0.0150 | No | ||
| 2 | OSR1 | 771 | 6.707 | -0.0136 | No | ||
| 3 | PCBP1 | 1141 | 5.320 | -0.0233 | No | ||
| 4 | GRSF1 | 1501 | 4.277 | -0.0346 | No | ||
| 5 | APRIN | 1509 | 4.258 | -0.0267 | No | ||
| 6 | CBX3 | 1542 | 4.181 | -0.0204 | No | ||
| 7 | LRRIQ2 | 1583 | 4.101 | -0.0146 | No | ||
| 8 | ACIN1 | 1819 | 3.618 | -0.0204 | No | ||
| 9 | BHLHB3 | 1908 | 3.447 | -0.0185 | No | ||
| 10 | MSH6 | 2162 | 3.058 | -0.0263 | No | ||
| 11 | ACTN4 | 2317 | 2.836 | -0.0292 | No | ||
| 12 | EGR3 | 2326 | 2.825 | -0.0242 | No | ||
| 13 | CRK | 2332 | 2.817 | -0.0190 | No | ||
| 14 | SPOCK2 | 2525 | 2.592 | -0.0244 | No | ||
| 15 | PPM1G | 2533 | 2.586 | -0.0198 | No | ||
| 16 | IL18BP | 2637 | 2.490 | -0.0205 | No | ||
| 17 | CAMK2A | 2783 | 2.365 | -0.0238 | No | ||
| 18 | SIX4 | 2897 | 2.266 | -0.0256 | No | ||
| 19 | HMGCLL1 | 2908 | 2.254 | -0.0218 | No | ||
| 20 | UNC84A | 2959 | 2.209 | -0.0202 | No | ||
| 21 | RHOT1 | 2982 | 2.194 | -0.0172 | No | ||
| 22 | ATG16L1 | 3025 | 2.150 | -0.0153 | No | ||
| 23 | ZFYVE21 | 3533 | 1.799 | -0.0394 | No | ||
| 24 | REV3L | 3642 | 1.740 | -0.0419 | No | ||
| 25 | ELAVL4 | 3646 | 1.739 | -0.0387 | No | ||
| 26 | NRG1 | 3661 | 1.731 | -0.0361 | No | ||
| 27 | SNX16 | 3702 | 1.706 | -0.0349 | No | ||
| 28 | SRF | 3711 | 1.702 | -0.0321 | No | ||
| 29 | HOXA13 | 3764 | 1.673 | -0.0317 | No | ||
| 30 | SCOC | 3849 | 1.628 | -0.0331 | No | ||
| 31 | RIPK5 | 3997 | 1.554 | -0.0381 | No | ||
| 32 | LMX1A | 4025 | 1.546 | -0.0365 | No | ||
| 33 | NPAT | 4274 | 1.439 | -0.0472 | No | ||
| 34 | SP2 | 4348 | 1.409 | -0.0485 | No | ||
| 35 | AHR | 4383 | 1.398 | -0.0476 | No | ||
| 36 | APBB3 | 4390 | 1.395 | -0.0452 | No | ||
| 37 | NTRK3 | 4428 | 1.382 | -0.0446 | No | ||
| 38 | MBD2 | 4660 | 1.289 | -0.0546 | No | ||
| 39 | STC1 | 4879 | 1.204 | -0.0641 | No | ||
| 40 | SPEN | 5044 | 1.158 | -0.0708 | No | ||
| 41 | SLC17A7 | 5069 | 1.151 | -0.0699 | No | ||
| 42 | COL4A3 | 5099 | 1.140 | -0.0692 | No | ||
| 43 | EXOC5 | 5339 | 1.070 | -0.0802 | No | ||
| 44 | CPEB3 | 5412 | 1.040 | -0.0821 | No | ||
| 45 | UCHL3 | 5571 | 0.992 | -0.0887 | No | ||
| 46 | MMP24 | 5835 | 0.915 | -0.1012 | No | ||
| 47 | ZCCHC14 | 5839 | 0.913 | -0.0996 | No | ||
| 48 | ADAMTS5 | 5908 | 0.895 | -0.1016 | No | ||
| 49 | CBLN4 | 6033 | 0.859 | -0.1067 | No | ||
| 50 | MYF5 | 6040 | 0.856 | -0.1053 | No | ||
| 51 | CAPN7 | 6069 | 0.848 | -0.1052 | No | ||
| 52 | HIAT1 | 6182 | 0.818 | -0.1097 | No | ||
| 53 | ADAMTS1 | 6338 | 0.778 | -0.1166 | No | ||
| 54 | PCTK2 | 6405 | 0.762 | -0.1187 | No | ||
| 55 | RHOC | 6523 | 0.734 | -0.1237 | No | ||
| 56 | MYO1D | 6707 | 0.684 | -0.1323 | No | ||
| 57 | GARNL1 | 6785 | 0.663 | -0.1352 | No | ||
| 58 | SLC4A4 | 6841 | 0.649 | -0.1369 | No | ||
| 59 | KLF11 | 6845 | 0.648 | -0.1358 | No | ||
| 60 | CRSP2 | 6901 | 0.632 | -0.1376 | No | ||
| 61 | STK35 | 6946 | 0.621 | -0.1388 | No | ||
| 62 | BECN1 | 7028 | 0.600 | -0.1420 | No | ||
| 63 | SON | 7053 | 0.595 | -0.1422 | No | ||
| 64 | LRP2 | 7283 | 0.541 | -0.1536 | No | ||
| 65 | IGF1 | 7424 | 0.506 | -0.1602 | No | ||
| 66 | FNDC3A | 7427 | 0.504 | -0.1593 | No | ||
| 67 | TAOK2 | 7462 | 0.494 | -0.1602 | No | ||
| 68 | VANGL1 | 7532 | 0.476 | -0.1630 | No | ||
| 69 | NFAT5 | 7549 | 0.473 | -0.1630 | No | ||
| 70 | WDR26 | 7550 | 0.473 | -0.1621 | No | ||
| 71 | RERG | 7728 | 0.423 | -0.1709 | No | ||
| 72 | DCHS1 | 7913 | 0.377 | -0.1801 | No | ||
| 73 | CNNM4 | 7983 | 0.361 | -0.1832 | No | ||
| 74 | TRIP10 | 8147 | 0.319 | -0.1914 | No | ||
| 75 | ST8SIA2 | 8199 | 0.304 | -0.1936 | No | ||
| 76 | MGAT4B | 8270 | 0.285 | -0.1969 | No | ||
| 77 | CPEB2 | 8319 | 0.273 | -0.1989 | No | ||
| 78 | RPS6KA4 | 8330 | 0.271 | -0.1990 | No | ||
| 79 | ELAVL2 | 8546 | 0.220 | -0.2102 | No | ||
| 80 | LUZP2 | 8592 | 0.209 | -0.2123 | No | ||
| 81 | PAIP1 | 8663 | 0.192 | -0.2157 | No | ||
| 82 | KIT | 8853 | 0.149 | -0.2257 | No | ||
| 83 | ZFPM2 | 8855 | 0.148 | -0.2254 | No | ||
| 84 | EYA4 | 8909 | 0.136 | -0.2281 | No | ||
| 85 | SHANK2 | 8938 | 0.129 | -0.2293 | No | ||
| 86 | FOS | 9018 | 0.107 | -0.2334 | No | ||
| 87 | TPBG | 9056 | 0.097 | -0.2352 | No | ||
| 88 | BVES | 9170 | 0.064 | -0.2412 | No | ||
| 89 | BAI3 | 9280 | 0.037 | -0.2471 | No | ||
| 90 | RAB6B | 9323 | 0.027 | -0.2493 | No | ||
| 91 | EPAS1 | 9487 | -0.019 | -0.2581 | No | ||
| 92 | PCBP2 | 9615 | -0.051 | -0.2649 | No | ||
| 93 | ABCA1 | 9699 | -0.072 | -0.2693 | No | ||
| 94 | LRP1B | 9879 | -0.120 | -0.2788 | No | ||
| 95 | UBE2D1 | 9942 | -0.138 | -0.2819 | No | ||
| 96 | GRM7 | 9951 | -0.140 | -0.2821 | No | ||
| 97 | BTBD7 | 9962 | -0.143 | -0.2823 | No | ||
| 98 | BMP2 | 10055 | -0.167 | -0.2870 | No | ||
| 99 | SLC18A2 | 10222 | -0.204 | -0.2956 | No | ||
| 100 | RHOQ | 10293 | -0.218 | -0.2990 | No | ||
| 101 | SREBF1 | 10461 | -0.251 | -0.3076 | No | ||
| 102 | PURA | 10543 | -0.271 | -0.3115 | No | ||
| 103 | FZD7 | 10570 | -0.278 | -0.3123 | No | ||
| 104 | GPR88 | 10733 | -0.316 | -0.3205 | No | ||
| 105 | FIGN | 10823 | -0.338 | -0.3247 | No | ||
| 106 | KLF10 | 10954 | -0.374 | -0.3311 | No | ||
| 107 | MASTL | 10959 | -0.375 | -0.3305 | No | ||
| 108 | LHX8 | 10985 | -0.382 | -0.3312 | No | ||
| 109 | ABCG4 | 11000 | -0.383 | -0.3312 | No | ||
| 110 | UBE3C | 11084 | -0.403 | -0.3349 | No | ||
| 111 | MGLL | 11143 | -0.420 | -0.3373 | No | ||
| 112 | ROBO1 | 11219 | -0.440 | -0.3405 | No | ||
| 113 | HSPA8 | 11388 | -0.482 | -0.3487 | No | ||
| 114 | BAZ2A | 11443 | -0.495 | -0.3506 | No | ||
| 115 | CCNT2 | 11544 | -0.518 | -0.3551 | No | ||
| 116 | CUL2 | 11578 | -0.529 | -0.3558 | No | ||
| 117 | RGMA | 11785 | -0.586 | -0.3659 | No | ||
| 118 | FXYD3 | 11839 | -0.603 | -0.3676 | No | ||
| 119 | HIPK1 | 11861 | -0.608 | -0.3676 | No | ||
| 120 | RTN1 | 12130 | -0.684 | -0.3808 | No | ||
| 121 | PTPN4 | 12190 | -0.699 | -0.3827 | No | ||
| 122 | RNF128 | 12334 | -0.746 | -0.3890 | No | ||
| 123 | DIO2 | 12477 | -0.792 | -0.3952 | No | ||
| 124 | UBE4A | 12504 | -0.802 | -0.3950 | No | ||
| 125 | SLC36A1 | 12548 | -0.814 | -0.3958 | No | ||
| 126 | VPS54 | 12569 | -0.822 | -0.3953 | No | ||
| 127 | PRPF4B | 12623 | -0.836 | -0.3965 | No | ||
| 128 | RAB11FIP5 | 12680 | -0.852 | -0.3979 | No | ||
| 129 | KCNJ3 | 12767 | -0.879 | -0.4009 | No | ||
| 130 | SLC25A27 | 13112 | -0.982 | -0.4177 | No | ||
| 131 | ULK1 | 13173 | -1.006 | -0.4190 | No | ||
| 132 | TRIM36 | 13397 | -1.087 | -0.4290 | No | ||
| 133 | PLEKHA3 | 13460 | -1.112 | -0.4302 | No | ||
| 134 | PAPOLB | 13543 | -1.146 | -0.4325 | No | ||
| 135 | ALS2 | 13557 | -1.152 | -0.4310 | No | ||
| 136 | RCN2 | 13682 | -1.203 | -0.4354 | No | ||
| 137 | OTX2 | 13853 | -1.269 | -0.4421 | No | ||
| 138 | STAG1 | 13906 | -1.284 | -0.4425 | No | ||
| 139 | TRIM3 | 13999 | -1.321 | -0.4449 | No | ||
| 140 | QKI | 14028 | -1.333 | -0.4439 | No | ||
| 141 | PAPPA | 14160 | -1.382 | -0.4483 | No | ||
| 142 | NFE2L2 | 14171 | -1.389 | -0.4462 | No | ||
| 143 | BTBD1 | 14195 | -1.397 | -0.4447 | No | ||
| 144 | CLCN3 | 14331 | -1.451 | -0.4492 | No | ||
| 145 | KITLG | 14610 | -1.576 | -0.4613 | No | ||
| 146 | CELSR1 | 14629 | -1.585 | -0.4592 | No | ||
| 147 | KCNRG | 14631 | -1.588 | -0.4562 | No | ||
| 148 | SYT7 | 14655 | -1.603 | -0.4543 | No | ||
| 149 | MEA1 | 14692 | -1.624 | -0.4531 | No | ||
| 150 | ALCAM | 14756 | -1.657 | -0.4534 | No | ||
| 151 | CDK5 | 14854 | -1.716 | -0.4553 | No | ||
| 152 | RPS25 | 14928 | -1.756 | -0.4559 | No | ||
| 153 | CHD9 | 15058 | -1.831 | -0.4593 | No | ||
| 154 | PBX3 | 15182 | -1.918 | -0.4623 | No | ||
| 155 | DDHD1 | 15260 | -1.982 | -0.4627 | No | ||
| 156 | FBXL3 | 15314 | -2.031 | -0.4616 | No | ||
| 157 | PLXNA2 | 15371 | -2.070 | -0.4606 | No | ||
| 158 | CUL4A | 15491 | -2.160 | -0.4629 | No | ||
| 159 | MBIP | 15516 | -2.175 | -0.4600 | No | ||
| 160 | JMJD1C | 15536 | -2.190 | -0.4568 | No | ||
| 161 | MED28 | 15590 | -2.239 | -0.4554 | No | ||
| 162 | UBR1 | 15665 | -2.306 | -0.4549 | No | ||
| 163 | FGF13 | 15966 | -2.612 | -0.4662 | Yes | ||
| 164 | ARID2 | 16040 | -2.705 | -0.4649 | Yes | ||
| 165 | MAT2B | 16070 | -2.742 | -0.4612 | Yes | ||
| 166 | RNH1 | 16153 | -2.846 | -0.4601 | Yes | ||
| 167 | NPAS4 | 16339 | -3.120 | -0.4641 | Yes | ||
| 168 | LRP12 | 16352 | -3.140 | -0.4587 | Yes | ||
| 169 | AP1G1 | 16381 | -3.175 | -0.4541 | Yes | ||
| 170 | TAF10 | 16402 | -3.220 | -0.4489 | Yes | ||
| 171 | FOXJ2 | 16451 | -3.297 | -0.4452 | Yes | ||
| 172 | ATXN7L2 | 16537 | -3.447 | -0.4431 | Yes | ||
| 173 | GOPC | 16621 | -3.596 | -0.4407 | Yes | ||
| 174 | HN1 | 16641 | -3.637 | -0.4347 | Yes | ||
| 175 | ZFX | 16712 | -3.751 | -0.4312 | Yes | ||
| 176 | BNIP2 | 16899 | -4.130 | -0.4333 | Yes | ||
| 177 | MKLN1 | 16977 | -4.324 | -0.4291 | Yes | ||
| 178 | H3F3A | 17152 | -4.820 | -0.4293 | Yes | ||
| 179 | SRI | 17247 | -5.079 | -0.4245 | Yes | ||
| 180 | CIC | 17278 | -5.164 | -0.4162 | Yes | ||
| 181 | TRPC4AP | 17286 | -5.185 | -0.4065 | Yes | ||
| 182 | ARID4B | 17348 | -5.382 | -0.3994 | Yes | ||
| 183 | ZCCHC11 | 17382 | -5.487 | -0.3906 | Yes | ||
| 184 | SDCBP | 17387 | -5.502 | -0.3802 | Yes | ||
| 185 | UBE2A | 17557 | -6.086 | -0.3776 | Yes | ||
| 186 | CD69 | 17611 | -6.271 | -0.3683 | Yes | ||
| 187 | HOXB4 | 17648 | -6.367 | -0.3580 | Yes | ||
| 188 | GAS7 | 17671 | -6.425 | -0.3467 | Yes | ||
| 189 | RHOA | 17735 | -6.687 | -0.3372 | Yes | ||
| 190 | TGFBR2 | 17742 | -6.708 | -0.3246 | Yes | ||
| 191 | RPS6KA5 | 17960 | -7.702 | -0.3215 | Yes | ||
| 192 | ETV1 | 17982 | -7.808 | -0.3075 | Yes | ||
| 193 | MCL1 | 18083 | -8.510 | -0.2965 | Yes | ||
| 194 | SSH2 | 18101 | -8.657 | -0.2806 | Yes | ||
| 195 | BTG3 | 18130 | -8.934 | -0.2649 | Yes | ||
| 196 | CCNG2 | 18163 | -9.168 | -0.2489 | Yes | ||
| 197 | DUSP2 | 18223 | -9.755 | -0.2332 | Yes | ||
| 198 | ATP1B1 | 18264 | -10.083 | -0.2159 | Yes | ||
| 199 | DNAJC7 | 18266 | -10.101 | -0.1964 | Yes | ||
| 200 | HERPUD1 | 18310 | -10.670 | -0.1781 | Yes | ||
| 201 | EGLN3 | 18317 | -10.740 | -0.1576 | Yes | ||
| 202 | TCF12 | 18338 | -11.172 | -0.1371 | Yes | ||
| 203 | ZFP36 | 18359 | -11.461 | -0.1160 | Yes | ||
| 204 | ZFAND3 | 18368 | -11.579 | -0.0940 | Yes | ||
| 205 | BTG1 | 18371 | -11.636 | -0.0716 | Yes | ||
| 206 | GCH1 | 18394 | -12.185 | -0.0492 | Yes | ||
| 207 | TLE4 | 18445 | -13.127 | -0.0266 | Yes | ||
| 208 | SOX5 | 18531 | -18.503 | 0.0046 | Yes |