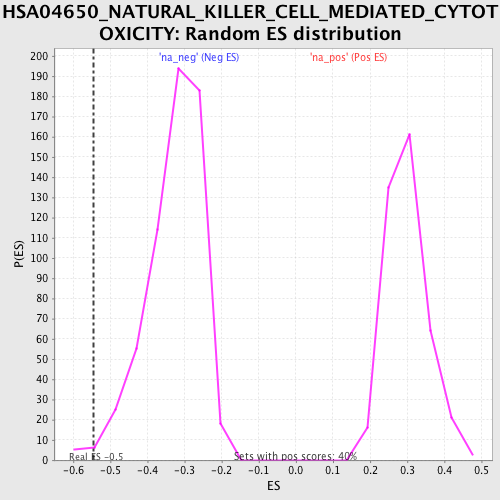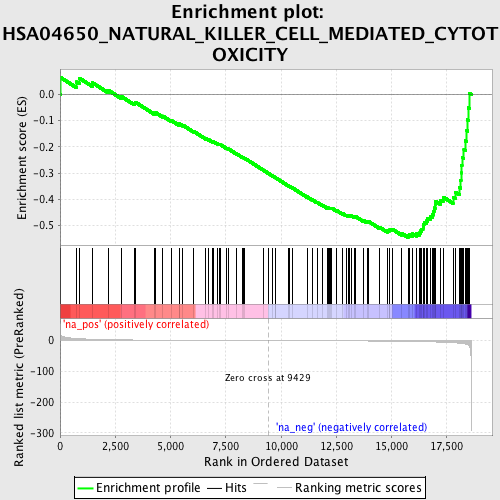
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA04650_NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY |
| Enrichment Score (ES) | -0.5448892 |
| Normalized Enrichment Score (NES) | -1.6630269 |
| Nominal p-value | 0.01 |
| FDR q-value | 0.5278856 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BID | 28 | 19.750 | 0.0643 | No | ||
| 2 | CD48 | 752 | 6.767 | 0.0478 | No | ||
| 3 | PIK3CB | 882 | 6.246 | 0.0616 | No | ||
| 4 | PIK3R3 | 1472 | 4.347 | 0.0443 | No | ||
| 5 | HCST | 2185 | 3.017 | 0.0159 | No | ||
| 6 | PPP3R2 | 2758 | 2.385 | -0.0070 | No | ||
| 7 | IFNA7 | 3369 | 1.898 | -0.0336 | No | ||
| 8 | MAP2K1 | 3408 | 1.872 | -0.0295 | No | ||
| 9 | FCER1G | 4292 | 1.432 | -0.0724 | No | ||
| 10 | IFNA14 | 4331 | 1.417 | -0.0697 | No | ||
| 11 | SH2D1B | 4641 | 1.300 | -0.0820 | No | ||
| 12 | IFNA1 | 5032 | 1.160 | -0.0992 | No | ||
| 13 | NCR1 | 5395 | 1.044 | -0.1153 | No | ||
| 14 | MAP2K2 | 5396 | 1.043 | -0.1118 | No | ||
| 15 | IFNA4 | 5556 | 0.996 | -0.1171 | No | ||
| 16 | PAK1 | 6045 | 0.855 | -0.1406 | No | ||
| 17 | SHC3 | 6599 | 0.710 | -0.1681 | No | ||
| 18 | PIK3R5 | 6715 | 0.682 | -0.1720 | No | ||
| 19 | IFNB1 | 6917 | 0.628 | -0.1808 | No | ||
| 20 | CD247 | 6934 | 0.624 | -0.1796 | No | ||
| 21 | RAC3 | 7111 | 0.581 | -0.1871 | No | ||
| 22 | CASP3 | 7210 | 0.556 | -0.1906 | No | ||
| 23 | KIR3DL1 | 7243 | 0.550 | -0.1905 | No | ||
| 24 | NFAT5 | 7549 | 0.473 | -0.2054 | No | ||
| 25 | KLRC1 | 7617 | 0.453 | -0.2075 | No | ||
| 26 | PIK3R2 | 7997 | 0.358 | -0.2267 | No | ||
| 27 | ITGAL | 8267 | 0.285 | -0.2403 | No | ||
| 28 | FCGR3A | 8278 | 0.282 | -0.2399 | No | ||
| 29 | PRF1 | 8338 | 0.269 | -0.2422 | No | ||
| 30 | HRAS | 9216 | 0.053 | -0.2894 | No | ||
| 31 | LAT | 9409 | 0.005 | -0.2997 | No | ||
| 32 | NRAS | 9596 | -0.046 | -0.3096 | No | ||
| 33 | PLCG1 | 9738 | -0.081 | -0.3170 | No | ||
| 34 | TNFSF10 | 9760 | -0.087 | -0.3178 | No | ||
| 35 | SOS1 | 10324 | -0.226 | -0.3474 | No | ||
| 36 | IFNAR1 | 10371 | -0.234 | -0.3492 | No | ||
| 37 | IFNA6 | 10510 | -0.262 | -0.3557 | No | ||
| 38 | ARAF | 11175 | -0.429 | -0.3902 | No | ||
| 39 | TYROBP | 11412 | -0.486 | -0.4013 | No | ||
| 40 | IFNA2 | 11632 | -0.543 | -0.4113 | No | ||
| 41 | PIK3R1 | 11865 | -0.608 | -0.4218 | No | ||
| 42 | KLRC2 | 12087 | -0.673 | -0.4315 | No | ||
| 43 | NFATC2 | 12146 | -0.688 | -0.4323 | No | ||
| 44 | KLRC3 | 12204 | -0.704 | -0.4331 | No | ||
| 45 | NFATC4 | 12252 | -0.716 | -0.4332 | No | ||
| 46 | TNF | 12300 | -0.734 | -0.4333 | No | ||
| 47 | IFNA13 | 12517 | -0.806 | -0.4423 | No | ||
| 48 | KLRK1 | 12794 | -0.887 | -0.4542 | No | ||
| 49 | GZMB | 12950 | -0.930 | -0.4595 | No | ||
| 50 | PTPN11 | 13063 | -0.961 | -0.4623 | No | ||
| 51 | CSF2 | 13096 | -0.975 | -0.4608 | No | ||
| 52 | KLRD1 | 13195 | -1.016 | -0.4627 | No | ||
| 53 | SH2D1A | 13309 | -1.055 | -0.4653 | No | ||
| 54 | NFATC3 | 13378 | -1.079 | -0.4654 | No | ||
| 55 | IFNGR1 | 13718 | -1.219 | -0.4796 | No | ||
| 56 | TNFRSF10B | 13895 | -1.280 | -0.4849 | No | ||
| 57 | PPP3CA | 13975 | -1.312 | -0.4848 | No | ||
| 58 | FASLG | 14445 | -1.500 | -0.5051 | No | ||
| 59 | RAF1 | 14806 | -1.685 | -0.5189 | No | ||
| 60 | SHC1 | 14889 | -1.736 | -0.5176 | No | ||
| 61 | RAC1 | 14926 | -1.756 | -0.5137 | No | ||
| 62 | VAV2 | 15039 | -1.820 | -0.5136 | No | ||
| 63 | LCP2 | 15453 | -2.130 | -0.5289 | No | ||
| 64 | PRKCB1 | 15751 | -2.383 | -0.5370 | Yes | ||
| 65 | IFNGR2 | 15806 | -2.439 | -0.5317 | Yes | ||
| 66 | ICAM1 | 15958 | -2.602 | -0.5312 | Yes | ||
| 67 | ZAP70 | 16117 | -2.799 | -0.5304 | Yes | ||
| 68 | ITGB2 | 16250 | -2.965 | -0.5277 | Yes | ||
| 69 | FYN | 16296 | -3.029 | -0.5200 | Yes | ||
| 70 | SOS2 | 16367 | -3.158 | -0.5133 | Yes | ||
| 71 | PIK3CA | 16426 | -3.246 | -0.5056 | Yes | ||
| 72 | IFNA5 | 16446 | -3.285 | -0.4957 | Yes | ||
| 73 | IFNG | 16475 | -3.329 | -0.4861 | Yes | ||
| 74 | PIK3CD | 16600 | -3.558 | -0.4810 | Yes | ||
| 75 | GRB2 | 16640 | -3.636 | -0.4709 | Yes | ||
| 76 | PPP3CB | 16752 | -3.824 | -0.4642 | Yes | ||
| 77 | SH3BP2 | 16850 | -4.017 | -0.4561 | Yes | ||
| 78 | PLCG2 | 16913 | -4.165 | -0.4455 | Yes | ||
| 79 | PPP3R1 | 16929 | -4.205 | -0.4323 | Yes | ||
| 80 | PPP3CC | 16975 | -4.313 | -0.4204 | Yes | ||
| 81 | LCK | 16984 | -4.337 | -0.4064 | Yes | ||
| 82 | FAS | 17226 | -5.006 | -0.4027 | Yes | ||
| 83 | MAPK3 | 17335 | -5.344 | -0.3907 | Yes | ||
| 84 | MAPK1 | 17787 | -6.868 | -0.3922 | Yes | ||
| 85 | ICAM2 | 17892 | -7.418 | -0.3731 | Yes | ||
| 86 | SYK | 18076 | -8.451 | -0.3549 | Yes | ||
| 87 | VAV1 | 18125 | -8.843 | -0.3280 | Yes | ||
| 88 | PIK3CG | 18147 | -9.020 | -0.2991 | Yes | ||
| 89 | PRKCA | 18153 | -9.054 | -0.2692 | Yes | ||
| 90 | IFNAR2 | 18192 | -9.427 | -0.2398 | Yes | ||
| 91 | NFATC1 | 18277 | -10.197 | -0.2104 | Yes | ||
| 92 | RAC2 | 18348 | -11.277 | -0.1766 | Yes | ||
| 93 | KRAS | 18407 | -12.449 | -0.1383 | Yes | ||
| 94 | PTPN6 | 18441 | -13.086 | -0.0965 | Yes | ||
| 95 | PTK2B | 18476 | -14.219 | -0.0509 | Yes | ||
| 96 | VAV3 | 18521 | -17.543 | 0.0051 | Yes |