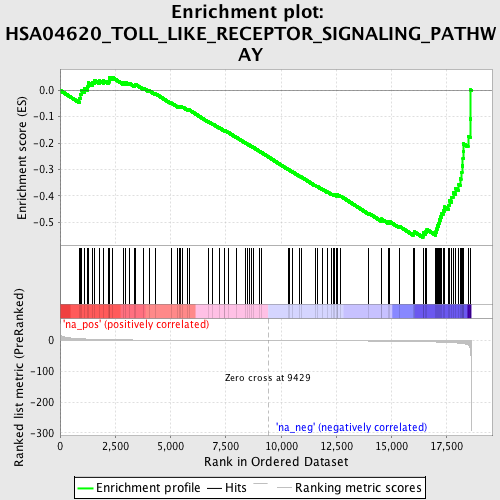
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA04620_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.5579313 |
| Normalized Enrichment Score (NES) | -1.6870089 |
| Nominal p-value | 0.020933978 |
| FDR q-value | 0.536762 |
| FWER p-Value | 0.998 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PIK3CB | 882 | 6.246 | -0.0295 | No | ||
| 2 | TIRAP | 928 | 6.071 | -0.0144 | No | ||
| 3 | MAP3K7 | 972 | 5.905 | 0.0004 | No | ||
| 4 | MAPK11 | 1109 | 5.409 | 0.0087 | No | ||
| 5 | CD86 | 1255 | 4.945 | 0.0151 | No | ||
| 6 | MAPK12 | 1269 | 4.916 | 0.0287 | No | ||
| 7 | PIK3R3 | 1472 | 4.347 | 0.0303 | No | ||
| 8 | IRAK1 | 1556 | 4.142 | 0.0378 | No | ||
| 9 | TICAM2 | 1771 | 3.735 | 0.0371 | No | ||
| 10 | IKBKB | 1959 | 3.365 | 0.0367 | No | ||
| 11 | MAPK8 | 2179 | 3.027 | 0.0337 | No | ||
| 12 | CHUK | 2221 | 2.966 | 0.0400 | No | ||
| 13 | TRAF3 | 2235 | 2.939 | 0.0478 | No | ||
| 14 | MAP3K7IP1 | 2375 | 2.758 | 0.0483 | No | ||
| 15 | NFKB2 | 2856 | 2.303 | 0.0291 | No | ||
| 16 | AKT1 | 2974 | 2.198 | 0.0291 | No | ||
| 17 | MYD88 | 3117 | 2.089 | 0.0275 | No | ||
| 18 | IFNA7 | 3369 | 1.898 | 0.0194 | No | ||
| 19 | MAP2K1 | 3408 | 1.872 | 0.0228 | No | ||
| 20 | TLR7 | 3761 | 1.674 | 0.0086 | No | ||
| 21 | CXCL9 | 4028 | 1.545 | -0.0013 | No | ||
| 22 | IFNA14 | 4331 | 1.417 | -0.0135 | No | ||
| 23 | IFNA1 | 5032 | 1.160 | -0.0479 | No | ||
| 24 | IL6 | 5329 | 1.073 | -0.0608 | No | ||
| 25 | MAP2K2 | 5396 | 1.043 | -0.0613 | No | ||
| 26 | CD14 | 5445 | 1.030 | -0.0609 | No | ||
| 27 | IFNA4 | 5556 | 0.996 | -0.0640 | No | ||
| 28 | IL1B | 5762 | 0.937 | -0.0724 | No | ||
| 29 | TLR8 | 5844 | 0.911 | -0.0741 | No | ||
| 30 | PIK3R5 | 6715 | 0.682 | -0.1191 | No | ||
| 31 | IFNB1 | 6917 | 0.628 | -0.1281 | No | ||
| 32 | CCL4 | 7202 | 0.559 | -0.1418 | No | ||
| 33 | CCL5 | 7441 | 0.500 | -0.1532 | No | ||
| 34 | TBK1 | 7451 | 0.497 | -0.1523 | No | ||
| 35 | MAPK13 | 7624 | 0.451 | -0.1603 | No | ||
| 36 | PIK3R2 | 7997 | 0.358 | -0.1793 | No | ||
| 37 | TLR5 | 8401 | 0.256 | -0.2003 | No | ||
| 38 | CCL3 | 8496 | 0.235 | -0.2047 | No | ||
| 39 | LBP | 8551 | 0.219 | -0.2070 | No | ||
| 40 | CXCL11 | 8643 | 0.198 | -0.2113 | No | ||
| 41 | IL12B | 8737 | 0.175 | -0.2159 | No | ||
| 42 | FOS | 9018 | 0.107 | -0.2307 | No | ||
| 43 | MAP3K8 | 9126 | 0.078 | -0.2362 | No | ||
| 44 | TLR3 | 10326 | -0.226 | -0.3003 | No | ||
| 45 | IFNAR1 | 10371 | -0.234 | -0.3020 | No | ||
| 46 | IFNA6 | 10510 | -0.262 | -0.3087 | No | ||
| 47 | MAP2K7 | 10828 | -0.340 | -0.3248 | No | ||
| 48 | IKBKE | 10915 | -0.364 | -0.3284 | No | ||
| 49 | TLR9 | 11539 | -0.517 | -0.3606 | No | ||
| 50 | IFNA2 | 11632 | -0.543 | -0.3639 | No | ||
| 51 | PIK3R1 | 11865 | -0.608 | -0.3747 | No | ||
| 52 | LY96 | 12097 | -0.675 | -0.3852 | No | ||
| 53 | TNF | 12300 | -0.734 | -0.3940 | No | ||
| 54 | IRF7 | 12392 | -0.765 | -0.3967 | No | ||
| 55 | RIPK1 | 12399 | -0.767 | -0.3948 | No | ||
| 56 | IFNA13 | 12517 | -0.806 | -0.3988 | No | ||
| 57 | CD80 | 12540 | -0.812 | -0.3976 | No | ||
| 58 | FADD | 12551 | -0.815 | -0.3958 | No | ||
| 59 | IRF5 | 12706 | -0.862 | -0.4016 | No | ||
| 60 | CXCL10 | 13979 | -1.314 | -0.4665 | No | ||
| 61 | MAP2K3 | 14531 | -1.538 | -0.4918 | No | ||
| 62 | MAPK14 | 14537 | -1.541 | -0.4876 | No | ||
| 63 | TLR4 | 14840 | -1.707 | -0.4990 | No | ||
| 64 | RAC1 | 14926 | -1.756 | -0.4985 | No | ||
| 65 | MAPK10 | 15352 | -2.059 | -0.5155 | No | ||
| 66 | CASP8 | 15978 | -2.634 | -0.5416 | No | ||
| 67 | MAP2K6 | 16017 | -2.684 | -0.5359 | No | ||
| 68 | PIK3CA | 16426 | -3.246 | -0.5485 | Yes | ||
| 69 | IFNA5 | 16446 | -3.285 | -0.5401 | Yes | ||
| 70 | IKBKG | 16553 | -3.477 | -0.5357 | Yes | ||
| 71 | PIK3CD | 16600 | -3.558 | -0.5279 | Yes | ||
| 72 | MAP2K4 | 17006 | -4.402 | -0.5370 | Yes | ||
| 73 | IL12A | 17038 | -4.515 | -0.5257 | Yes | ||
| 74 | RELA | 17065 | -4.586 | -0.5138 | Yes | ||
| 75 | JUN | 17135 | -4.762 | -0.5037 | Yes | ||
| 76 | IRAK4 | 17160 | -4.845 | -0.4910 | Yes | ||
| 77 | IRF3 | 17225 | -5.005 | -0.4800 | Yes | ||
| 78 | MAP3K7IP2 | 17275 | -5.161 | -0.4677 | Yes | ||
| 79 | MAPK3 | 17335 | -5.344 | -0.4554 | Yes | ||
| 80 | NFKB1 | 17383 | -5.487 | -0.4421 | Yes | ||
| 81 | MAPK9 | 17593 | -6.221 | -0.4354 | Yes | ||
| 82 | NFKBIA | 17625 | -6.306 | -0.4188 | Yes | ||
| 83 | TOLLIP | 17725 | -6.647 | -0.4049 | Yes | ||
| 84 | MAPK1 | 17787 | -6.868 | -0.3883 | Yes | ||
| 85 | CD40 | 17901 | -7.482 | -0.3728 | Yes | ||
| 86 | AKT2 | 18046 | -8.193 | -0.3568 | Yes | ||
| 87 | AKT3 | 18107 | -8.687 | -0.3350 | Yes | ||
| 88 | PIK3CG | 18147 | -9.020 | -0.3110 | Yes | ||
| 89 | IFNAR2 | 18192 | -9.427 | -0.2861 | Yes | ||
| 90 | TRAF6 | 18232 | -9.851 | -0.2597 | Yes | ||
| 91 | STAT1 | 18246 | -9.978 | -0.2315 | Yes | ||
| 92 | TLR2 | 18257 | -10.040 | -0.2030 | Yes | ||
| 93 | TLR1 | 18465 | -13.850 | -0.1741 | Yes | ||
| 94 | SPP1 | 18561 | -24.511 | -0.1083 | Yes | ||
| 95 | TLR6 | 18585 | -38.435 | 0.0017 | Yes |
