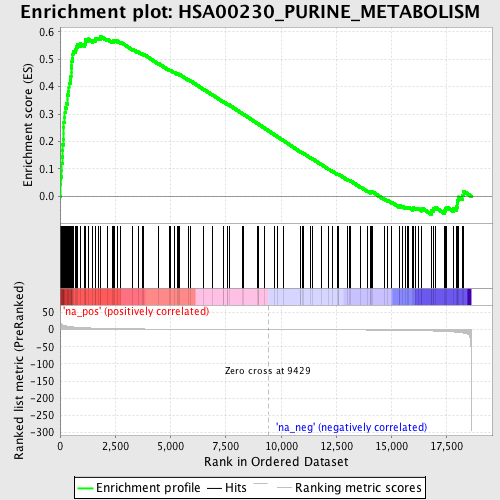
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HSA00230_PURINE_METABOLISM |
| Enrichment Score (ES) | 0.5843934 |
| Normalized Enrichment Score (NES) | 2.0713422 |
| Nominal p-value | 0.0 |
| FDR q-value | 3.7017427E-4 |
| FWER p-Value | 0.0030 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | POLR1B | 13 | 25.660 | 0.0431 | Yes | ||
| 2 | IMPDH2 | 40 | 17.301 | 0.0712 | Yes | ||
| 3 | NME2 | 80 | 14.966 | 0.0946 | Yes | ||
| 4 | NME1 | 84 | 14.861 | 0.1198 | Yes | ||
| 5 | ADK | 94 | 14.400 | 0.1439 | Yes | ||
| 6 | PPAT | 107 | 13.796 | 0.1668 | Yes | ||
| 7 | POLR1A | 117 | 13.455 | 0.1893 | Yes | ||
| 8 | POLD2 | 156 | 12.471 | 0.2085 | Yes | ||
| 9 | GART | 162 | 12.281 | 0.2292 | Yes | ||
| 10 | AK2 | 163 | 12.258 | 0.2501 | Yes | ||
| 11 | POLE | 169 | 12.131 | 0.2706 | Yes | ||
| 12 | APRT | 214 | 11.235 | 0.2874 | Yes | ||
| 13 | ADSL | 220 | 11.140 | 0.3061 | Yes | ||
| 14 | POLE3 | 248 | 10.705 | 0.3229 | Yes | ||
| 15 | ADA | 282 | 10.360 | 0.3388 | Yes | ||
| 16 | PKM2 | 328 | 9.829 | 0.3532 | Yes | ||
| 17 | POLR3H | 329 | 9.824 | 0.3699 | Yes | ||
| 18 | AMPD2 | 382 | 9.297 | 0.3830 | Yes | ||
| 19 | ATIC | 400 | 9.179 | 0.3977 | Yes | ||
| 20 | NME6 | 403 | 9.141 | 0.4132 | Yes | ||
| 21 | NME4 | 458 | 8.678 | 0.4251 | Yes | ||
| 22 | NUDT5 | 485 | 8.446 | 0.4381 | Yes | ||
| 23 | GMPS | 496 | 8.376 | 0.4519 | Yes | ||
| 24 | NUDT9 | 499 | 8.341 | 0.4660 | Yes | ||
| 25 | RFC5 | 526 | 8.111 | 0.4784 | Yes | ||
| 26 | POLA2 | 532 | 8.055 | 0.4919 | Yes | ||
| 27 | POLD3 | 550 | 7.954 | 0.5046 | Yes | ||
| 28 | POLR2J | 575 | 7.790 | 0.5166 | Yes | ||
| 29 | GUK1 | 586 | 7.684 | 0.5291 | Yes | ||
| 30 | PAICS | 703 | 6.999 | 0.5348 | Yes | ||
| 31 | ENTPD4 | 723 | 6.900 | 0.5455 | Yes | ||
| 32 | POLR2H | 779 | 6.679 | 0.5540 | Yes | ||
| 33 | NT5E | 917 | 6.120 | 0.5570 | Yes | ||
| 34 | NME7 | 1116 | 5.394 | 0.5555 | Yes | ||
| 35 | RRM1 | 1140 | 5.328 | 0.5633 | Yes | ||
| 36 | POLR2E | 1154 | 5.265 | 0.5716 | Yes | ||
| 37 | NUDT2 | 1265 | 4.922 | 0.5741 | Yes | ||
| 38 | HPRT1 | 1487 | 4.299 | 0.5695 | Yes | ||
| 39 | PDE8A | 1584 | 4.096 | 0.5713 | Yes | ||
| 40 | POLR3A | 1611 | 4.041 | 0.5767 | Yes | ||
| 41 | PRIM1 | 1730 | 3.806 | 0.5769 | Yes | ||
| 42 | POLR2A | 1817 | 3.619 | 0.5784 | Yes | ||
| 43 | PRPS1 | 1821 | 3.615 | 0.5844 | Yes | ||
| 44 | POLR1D | 2137 | 3.094 | 0.5726 | No | ||
| 45 | ALLC | 2367 | 2.769 | 0.5650 | No | ||
| 46 | ENTPD6 | 2415 | 2.702 | 0.5670 | No | ||
| 47 | ECGF1 | 2450 | 2.669 | 0.5698 | No | ||
| 48 | PNPT1 | 2584 | 2.537 | 0.5669 | No | ||
| 49 | PRPS2 | 2741 | 2.402 | 0.5626 | No | ||
| 50 | ADCY4 | 3289 | 1.959 | 0.5363 | No | ||
| 51 | POLR2K | 3525 | 1.806 | 0.5267 | No | ||
| 52 | AK5 | 3715 | 1.699 | 0.5194 | No | ||
| 53 | PDE10A | 3795 | 1.656 | 0.5179 | No | ||
| 54 | POLR2I | 4456 | 1.372 | 0.4846 | No | ||
| 55 | AK3L1 | 4952 | 1.179 | 0.4598 | No | ||
| 56 | ADCY3 | 4978 | 1.172 | 0.4604 | No | ||
| 57 | GDA | 5162 | 1.120 | 0.4525 | No | ||
| 58 | PAPSS2 | 5321 | 1.076 | 0.4457 | No | ||
| 59 | ITPA | 5342 | 1.069 | 0.4465 | No | ||
| 60 | ENTPD8 | 5421 | 1.037 | 0.4440 | No | ||
| 61 | GUCY2F | 5796 | 0.927 | 0.4254 | No | ||
| 62 | RRM2B | 5910 | 0.894 | 0.4208 | No | ||
| 63 | ENTPD3 | 6511 | 0.737 | 0.3896 | No | ||
| 64 | POLE2 | 6876 | 0.639 | 0.3710 | No | ||
| 65 | ENTPD1 | 7391 | 0.516 | 0.3441 | No | ||
| 66 | PDE6G | 7393 | 0.516 | 0.3449 | No | ||
| 67 | GUCY1A2 | 7573 | 0.468 | 0.3360 | No | ||
| 68 | XDH | 7579 | 0.466 | 0.3366 | No | ||
| 69 | AMPD1 | 7645 | 0.446 | 0.3338 | No | ||
| 70 | POLR2C | 8238 | 0.292 | 0.3023 | No | ||
| 71 | ADCY8 | 8284 | 0.281 | 0.3003 | No | ||
| 72 | PDE5A | 8918 | 0.133 | 0.2663 | No | ||
| 73 | NPR1 | 8969 | 0.120 | 0.2638 | No | ||
| 74 | PDE2A | 8981 | 0.117 | 0.2634 | No | ||
| 75 | POLR3B | 9239 | 0.048 | 0.2496 | No | ||
| 76 | POLA1 | 9723 | -0.077 | 0.2236 | No | ||
| 77 | NP | 9843 | -0.112 | 0.2174 | No | ||
| 78 | CANT1 | 10095 | -0.177 | 0.2041 | No | ||
| 79 | NPR2 | 10879 | -0.353 | 0.1623 | No | ||
| 80 | PDE1A | 10898 | -0.358 | 0.1620 | No | ||
| 81 | GUCY2C | 10979 | -0.381 | 0.1583 | No | ||
| 82 | PKLR | 10999 | -0.383 | 0.1579 | No | ||
| 83 | PDE3B | 11336 | -0.469 | 0.1406 | No | ||
| 84 | GUCY1B3 | 11433 | -0.492 | 0.1362 | No | ||
| 85 | PDE6H | 11832 | -0.601 | 0.1157 | No | ||
| 86 | ENTPD2 | 12153 | -0.690 | 0.0996 | No | ||
| 87 | PDE1C | 12339 | -0.747 | 0.0908 | No | ||
| 88 | PDE8B | 12542 | -0.812 | 0.0813 | No | ||
| 89 | AK1 | 12586 | -0.827 | 0.0804 | No | ||
| 90 | ENPP3 | 12590 | -0.827 | 0.0816 | No | ||
| 91 | POLR2G | 12990 | -0.941 | 0.0617 | No | ||
| 92 | ADCY6 | 13085 | -0.972 | 0.0582 | No | ||
| 93 | ENPP1 | 13138 | -0.993 | 0.0571 | No | ||
| 94 | PDE7B | 13607 | -1.175 | 0.0338 | No | ||
| 95 | ADCY5 | 13922 | -1.293 | 0.0190 | No | ||
| 96 | GUCY1A3 | 14050 | -1.339 | 0.0144 | No | ||
| 97 | PDE9A | 14063 | -1.345 | 0.0161 | No | ||
| 98 | NT5C3 | 14103 | -1.363 | 0.0163 | No | ||
| 99 | NT5C1B | 14120 | -1.369 | 0.0178 | No | ||
| 100 | POLR2B | 14675 | -1.616 | -0.0094 | No | ||
| 101 | PDE4D | 14829 | -1.703 | -0.0148 | No | ||
| 102 | NT5C | 15011 | -1.805 | -0.0215 | No | ||
| 103 | IMPDH1 | 15354 | -2.060 | -0.0365 | No | ||
| 104 | PDE4B | 15360 | -2.065 | -0.0332 | No | ||
| 105 | ADCY2 | 15475 | -2.150 | -0.0357 | No | ||
| 106 | POLR3K | 15639 | -2.284 | -0.0407 | No | ||
| 107 | PDE11A | 15727 | -2.364 | -0.0413 | No | ||
| 108 | PDE4A | 15759 | -2.395 | -0.0389 | No | ||
| 109 | AMPD3 | 15964 | -2.611 | -0.0455 | No | ||
| 110 | GMPR | 15976 | -2.634 | -0.0416 | No | ||
| 111 | FHIT | 16098 | -2.775 | -0.0434 | No | ||
| 112 | PDE4C | 16213 | -2.922 | -0.0446 | No | ||
| 113 | POLE4 | 16376 | -3.172 | -0.0479 | No | ||
| 114 | ADCY9 | 16377 | -3.172 | -0.0425 | No | ||
| 115 | PAPSS1 | 16811 | -3.942 | -0.0592 | No | ||
| 116 | POLD1 | 16812 | -3.942 | -0.0525 | No | ||
| 117 | NT5M | 16885 | -4.101 | -0.0494 | No | ||
| 118 | GMPR2 | 16887 | -4.103 | -0.0424 | No | ||
| 119 | ZNRD1 | 17008 | -4.403 | -0.0414 | No | ||
| 120 | ADCY7 | 17389 | -5.510 | -0.0526 | No | ||
| 121 | RRM2 | 17427 | -5.575 | -0.0450 | No | ||
| 122 | ENTPD5 | 17504 | -5.927 | -0.0390 | No | ||
| 123 | NT5C2 | 17807 | -6.965 | -0.0435 | No | ||
| 124 | ADSSL1 | 17927 | -7.609 | -0.0369 | No | ||
| 125 | DGUOK | 17976 | -7.781 | -0.0263 | No | ||
| 126 | DCK | 17991 | -7.855 | -0.0136 | No | ||
| 127 | PDE7A | 18037 | -8.127 | -0.0022 | No | ||
| 128 | POLD4 | 18204 | -9.516 | 0.0051 | No | ||
| 129 | PDE6D | 18258 | -10.042 | 0.0194 | No |