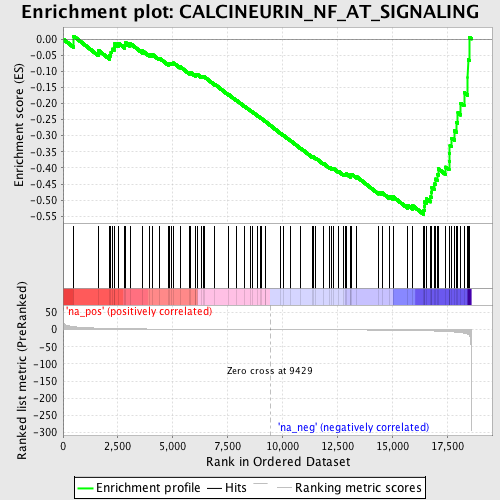
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CALCINEURIN_NF_AT_SIGNALING |
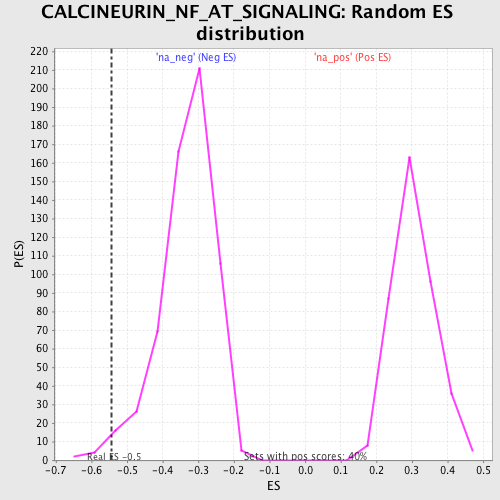
| Enrichment Score (ES) | -0.54446375 |
| Normalized Enrichment Score (NES) | -1.6444438 |
| Nominal p-value | 0.016528925 |
| FDR q-value | 0.48293966 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | XPO5 | 481 | 8.469 | 0.0090 | No | ||
| 2 | BCL2 | 1621 | 4.010 | -0.0359 | No | ||
| 3 | GATA4 | 2122 | 3.119 | -0.0500 | No | ||
| 4 | MAPK8 | 2179 | 3.027 | -0.0405 | No | ||
| 5 | CSNK2A1 | 2228 | 2.950 | -0.0309 | No | ||
| 6 | EGR3 | 2326 | 2.825 | -0.0245 | No | ||
| 7 | CEBPB | 2357 | 2.784 | -0.0146 | No | ||
| 8 | CNR1 | 2531 | 2.588 | -0.0133 | No | ||
| 9 | IL10 | 2814 | 2.335 | -0.0189 | No | ||
| 10 | NFKB2 | 2856 | 2.303 | -0.0116 | No | ||
| 11 | IL4 | 3049 | 2.137 | -0.0131 | No | ||
| 12 | PPIA | 3621 | 1.749 | -0.0367 | No | ||
| 13 | P2RX7 | 3933 | 1.590 | -0.0469 | No | ||
| 14 | ITK | 4064 | 1.527 | -0.0476 | No | ||
| 15 | TRPV6 | 4384 | 1.397 | -0.0591 | No | ||
| 16 | CSNK2B | 4799 | 1.233 | -0.0763 | No | ||
| 17 | RPL13A | 4865 | 1.207 | -0.0748 | No | ||
| 18 | CABIN1 | 4951 | 1.179 | -0.0745 | No | ||
| 19 | IFNA1 | 5032 | 1.160 | -0.0741 | No | ||
| 20 | IL6 | 5329 | 1.073 | -0.0856 | No | ||
| 21 | IL1B | 5762 | 0.937 | -0.1051 | No | ||
| 22 | IL3 | 5817 | 0.921 | -0.1042 | No | ||
| 23 | MYF5 | 6040 | 0.856 | -0.1126 | No | ||
| 24 | PAK1 | 6045 | 0.855 | -0.1093 | No | ||
| 25 | CALM1 | 6121 | 0.834 | -0.1099 | No | ||
| 26 | IL8RA | 6305 | 0.786 | -0.1165 | No | ||
| 27 | ACTB | 6384 | 0.769 | -0.1176 | No | ||
| 28 | ICOS | 6428 | 0.756 | -0.1168 | No | ||
| 29 | IFNB1 | 6917 | 0.628 | -0.1405 | No | ||
| 30 | NFAT5 | 7549 | 0.473 | -0.1726 | No | ||
| 31 | FCER1A | 7896 | 0.382 | -0.1897 | No | ||
| 32 | FCGR3A | 8278 | 0.282 | -0.2091 | No | ||
| 33 | CD3E | 8523 | 0.227 | -0.2213 | No | ||
| 34 | CD3G | 8635 | 0.200 | -0.2265 | No | ||
| 35 | IL13 | 8866 | 0.145 | -0.2383 | No | ||
| 36 | FOS | 9018 | 0.107 | -0.2460 | No | ||
| 37 | FOSL1 | 9050 | 0.097 | -0.2473 | No | ||
| 38 | HRAS | 9216 | 0.053 | -0.2560 | No | ||
| 39 | CAMK2B | 9238 | 0.048 | -0.2569 | No | ||
| 40 | TRAF2 | 9913 | -0.132 | -0.2928 | No | ||
| 41 | OPRD1 | 10062 | -0.168 | -0.3000 | No | ||
| 42 | CAMK4 | 10363 | -0.234 | -0.3153 | No | ||
| 43 | MAP2K7 | 10828 | -0.340 | -0.3389 | No | ||
| 44 | FKBP1B | 11378 | -0.480 | -0.3666 | No | ||
| 45 | NPPB | 11397 | -0.483 | -0.3655 | No | ||
| 46 | NFKBIB | 11524 | -0.516 | -0.3702 | No | ||
| 47 | GATA3 | 11864 | -0.608 | -0.3860 | No | ||
| 48 | NFATC2 | 12146 | -0.688 | -0.3983 | No | ||
| 49 | NFATC4 | 12252 | -0.716 | -0.4010 | No | ||
| 50 | TNF | 12300 | -0.734 | -0.4005 | No | ||
| 51 | SP1 | 12546 | -0.813 | -0.4104 | No | ||
| 52 | CTLA4 | 12772 | -0.881 | -0.4189 | No | ||
| 53 | CALM2 | 12857 | -0.904 | -0.4197 | No | ||
| 54 | SP3 | 12898 | -0.916 | -0.4181 | No | ||
| 55 | CSF2 | 13096 | -0.975 | -0.4247 | No | ||
| 56 | PIN1 | 13113 | -0.982 | -0.4215 | No | ||
| 57 | SFN | 13162 | -1.003 | -0.4199 | No | ||
| 58 | NFATC3 | 13378 | -1.079 | -0.4271 | No | ||
| 59 | GSK3A | 14380 | -1.471 | -0.4750 | No | ||
| 60 | MAPK14 | 14537 | -1.541 | -0.4771 | No | ||
| 61 | IL2 | 14885 | -1.734 | -0.4886 | No | ||
| 62 | VAV2 | 15039 | -1.820 | -0.4894 | No | ||
| 63 | MEF2D | 15700 | -2.341 | -0.5153 | No | ||
| 64 | NCK2 | 15934 | -2.580 | -0.5173 | No | ||
| 65 | PTPRC | 16439 | -3.278 | -0.5309 | Yes | ||
| 66 | JUNB | 16464 | -3.310 | -0.5186 | Yes | ||
| 67 | IFNG | 16475 | -3.329 | -0.5053 | Yes | ||
| 68 | CALM3 | 16568 | -3.499 | -0.4959 | Yes | ||
| 69 | PPP3CB | 16752 | -3.824 | -0.4899 | Yes | ||
| 70 | CREBBP | 16786 | -3.900 | -0.4756 | Yes | ||
| 71 | MEF2B | 16805 | -3.929 | -0.4604 | Yes | ||
| 72 | PPP3R1 | 16929 | -4.205 | -0.4496 | Yes | ||
| 73 | PPP3CC | 16975 | -4.313 | -0.4343 | Yes | ||
| 74 | RELA | 17065 | -4.586 | -0.4201 | Yes | ||
| 75 | BAD | 17107 | -4.710 | -0.4029 | Yes | ||
| 76 | GSK3B | 17428 | -5.589 | -0.3971 | Yes | ||
| 77 | MAPK9 | 17593 | -6.221 | -0.3802 | Yes | ||
| 78 | CD69 | 17611 | -6.271 | -0.3553 | Yes | ||
| 79 | IL2RA | 17631 | -6.321 | -0.3302 | Yes | ||
| 80 | TGFB1 | 17710 | -6.583 | -0.3072 | Yes | ||
| 81 | NFKBIE | 17840 | -7.112 | -0.2848 | Yes | ||
| 82 | MEF2A | 17935 | -7.632 | -0.2584 | Yes | ||
| 83 | CDKN1A | 17995 | -7.882 | -0.2290 | Yes | ||
| 84 | VAV1 | 18125 | -8.843 | -0.1994 | Yes | ||
| 85 | NFATC1 | 18277 | -10.197 | -0.1655 | Yes | ||
| 86 | SLA | 18451 | -13.415 | -0.1194 | Yes | ||
| 87 | EGR2 | 18453 | -13.500 | -0.0637 | Yes | ||
| 88 | VAV3 | 18521 | -17.543 | 0.0051 | Yes |