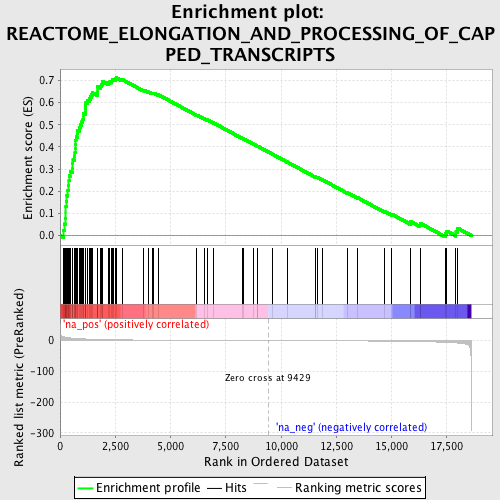
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | REACTOME_ELONGATION_AND_PROCESSING_OF_CAPPED_TRANSCRIPTS |
| Enrichment Score (ES) | 0.712696 |
| Normalized Enrichment Score (NES) | 2.3289545 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SNRPF | 151 | 12.568 | 0.0241 | Yes | ||
| 2 | U2AF1 | 184 | 11.817 | 0.0527 | Yes | ||
| 3 | SNRPD3 | 236 | 10.897 | 0.0779 | Yes | ||
| 4 | CPSF1 | 260 | 10.576 | 0.1038 | Yes | ||
| 5 | SFRS6 | 261 | 10.573 | 0.1309 | Yes | ||
| 6 | GTF2F2 | 284 | 10.329 | 0.1562 | Yes | ||
| 7 | EFTUD2 | 306 | 10.035 | 0.1808 | Yes | ||
| 8 | PHF5A | 351 | 9.518 | 0.2029 | Yes | ||
| 9 | THOC4 | 364 | 9.426 | 0.2264 | Yes | ||
| 10 | TXNL4A | 401 | 9.146 | 0.2479 | Yes | ||
| 11 | SNRPA1 | 424 | 8.961 | 0.2697 | Yes | ||
| 12 | CSTF3 | 453 | 8.709 | 0.2906 | Yes | ||
| 13 | GTF2H3 | 569 | 7.818 | 0.3044 | Yes | ||
| 14 | POLR2J | 575 | 7.790 | 0.3241 | Yes | ||
| 15 | DHX9 | 582 | 7.735 | 0.3436 | Yes | ||
| 16 | RNPS1 | 641 | 7.339 | 0.3593 | Yes | ||
| 17 | GTF2H1 | 671 | 7.198 | 0.3762 | Yes | ||
| 18 | YBX1 | 681 | 7.114 | 0.3940 | Yes | ||
| 19 | SF3A3 | 689 | 7.079 | 0.4118 | Yes | ||
| 20 | RDBP | 699 | 7.018 | 0.4293 | Yes | ||
| 21 | SF3B3 | 721 | 6.919 | 0.4459 | Yes | ||
| 22 | POLR2H | 779 | 6.679 | 0.4600 | Yes | ||
| 23 | PRPF4 | 802 | 6.608 | 0.4757 | Yes | ||
| 24 | NCBP2 | 887 | 6.207 | 0.4871 | Yes | ||
| 25 | SFRS5 | 921 | 6.090 | 0.5010 | Yes | ||
| 26 | SNRPG | 986 | 5.868 | 0.5126 | Yes | ||
| 27 | PRPF8 | 1032 | 5.686 | 0.5247 | Yes | ||
| 28 | GTF2F1 | 1058 | 5.591 | 0.5377 | Yes | ||
| 29 | SNRPD2 | 1061 | 5.582 | 0.5519 | Yes | ||
| 30 | PTBP1 | 1135 | 5.338 | 0.5617 | Yes | ||
| 31 | PCBP1 | 1141 | 5.320 | 0.5751 | Yes | ||
| 32 | POLR2E | 1154 | 5.265 | 0.5879 | Yes | ||
| 33 | SF3A1 | 1167 | 5.221 | 0.6007 | Yes | ||
| 34 | FUS | 1230 | 5.027 | 0.6102 | Yes | ||
| 35 | DNAJC8 | 1320 | 4.783 | 0.6177 | Yes | ||
| 36 | MAGOH | 1375 | 4.593 | 0.6266 | Yes | ||
| 37 | PABPN1 | 1434 | 4.430 | 0.6348 | Yes | ||
| 38 | SNRPA | 1459 | 4.368 | 0.6447 | Yes | ||
| 39 | SMC1A | 1682 | 3.894 | 0.6427 | Yes | ||
| 40 | SF3B2 | 1697 | 3.869 | 0.6519 | Yes | ||
| 41 | CSTF1 | 1701 | 3.859 | 0.6616 | Yes | ||
| 42 | SNRPB | 1703 | 3.855 | 0.6715 | Yes | ||
| 43 | POLR2A | 1817 | 3.619 | 0.6747 | Yes | ||
| 44 | SFRS2 | 1866 | 3.517 | 0.6811 | Yes | ||
| 45 | NFX1 | 1903 | 3.450 | 0.6880 | Yes | ||
| 46 | GTF2H2 | 1916 | 3.427 | 0.6961 | Yes | ||
| 47 | SFRS1 | 2175 | 3.032 | 0.6900 | Yes | ||
| 48 | CCAR1 | 2254 | 2.910 | 0.6933 | Yes | ||
| 49 | CSTF2 | 2347 | 2.796 | 0.6955 | Yes | ||
| 50 | SFRS9 | 2349 | 2.793 | 0.7026 | Yes | ||
| 51 | PAPOLA | 2417 | 2.701 | 0.7059 | Yes | ||
| 52 | MNAT1 | 2523 | 2.593 | 0.7069 | Yes | ||
| 53 | SFRS3 | 2539 | 2.582 | 0.7127 | Yes | ||
| 54 | TH1L | 2821 | 2.331 | 0.7035 | No | ||
| 55 | CCNH | 3774 | 1.669 | 0.6564 | No | ||
| 56 | TCEB2 | 4007 | 1.551 | 0.6479 | No | ||
| 57 | U2AF2 | 4166 | 1.488 | 0.6432 | No | ||
| 58 | NUDT21 | 4245 | 1.454 | 0.6427 | No | ||
| 59 | POLR2I | 4456 | 1.372 | 0.6349 | No | ||
| 60 | SF3A2 | 6186 | 0.816 | 0.5437 | No | ||
| 61 | PCF11 | 6527 | 0.731 | 0.5272 | No | ||
| 62 | ERCC3 | 6652 | 0.699 | 0.5223 | No | ||
| 63 | SFRS7 | 6937 | 0.624 | 0.5086 | No | ||
| 64 | POLR2C | 8238 | 0.292 | 0.4392 | No | ||
| 65 | SNRPE | 8301 | 0.278 | 0.4365 | No | ||
| 66 | TCEB1 | 8771 | 0.168 | 0.4117 | No | ||
| 67 | SF3B4 | 8944 | 0.128 | 0.4027 | No | ||
| 68 | SF4 | 8950 | 0.125 | 0.4028 | No | ||
| 69 | PCBP2 | 9615 | -0.051 | 0.3671 | No | ||
| 70 | CDK7 | 10270 | -0.214 | 0.3323 | No | ||
| 71 | CCNT2 | 11544 | -0.518 | 0.2650 | No | ||
| 72 | ELL | 11545 | -0.518 | 0.2663 | No | ||
| 73 | SNRPB2 | 11642 | -0.546 | 0.2625 | No | ||
| 74 | SRRM1 | 11871 | -0.610 | 0.2518 | No | ||
| 75 | POLR2G | 12990 | -0.941 | 0.1938 | No | ||
| 76 | METTL3 | 13472 | -1.116 | 0.1708 | No | ||
| 77 | POLR2B | 14675 | -1.616 | 0.1100 | No | ||
| 78 | SF3B1 | 15019 | -1.809 | 0.0962 | No | ||
| 79 | RBM5 | 15857 | -2.489 | 0.0574 | No | ||
| 80 | CCNT1 | 15870 | -2.503 | 0.0632 | No | ||
| 81 | SUPT4H1 | 16288 | -3.011 | 0.0484 | No | ||
| 82 | CD2BP2 | 16307 | -3.048 | 0.0552 | No | ||
| 83 | CDK9 | 17439 | -5.654 | 0.0087 | No | ||
| 84 | RBMX | 17495 | -5.879 | 0.0208 | No | ||
| 85 | TCEB3 | 17917 | -7.568 | 0.0175 | No | ||
| 86 | ERCC2 | 17992 | -7.856 | 0.0337 | No |
