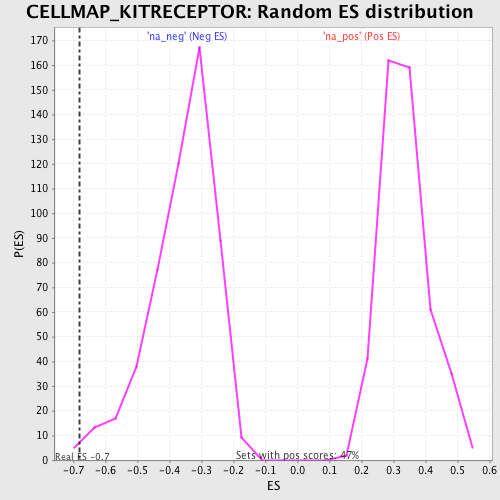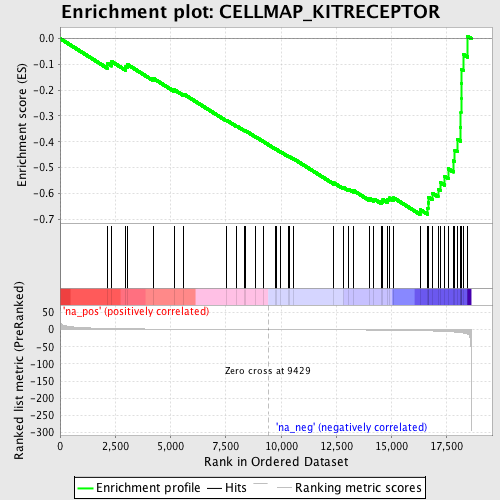
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CELLMAP_KITRECEPTOR |
| Enrichment Score (ES) | -0.68187875 |
| Normalized Enrichment Score (NES) | -1.8896526 |
| Nominal p-value | 0.0056074765 |
| FDR q-value | 0.69139916 |
| FWER p-Value | 0.939 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GRB7 | 2157 | 3.067 | -0.0971 | No | ||
| 2 | CRK | 2332 | 2.817 | -0.0889 | No | ||
| 3 | AKT1 | 2974 | 2.198 | -0.1098 | No | ||
| 4 | RPS6KA1 | 3055 | 2.134 | -0.1008 | No | ||
| 5 | KHDRBS1 | 4221 | 1.463 | -0.1545 | No | ||
| 6 | SRC | 5173 | 1.116 | -0.1987 | No | ||
| 7 | SPRED2 | 5595 | 0.986 | -0.2153 | No | ||
| 8 | CLTC | 7528 | 0.477 | -0.3163 | No | ||
| 9 | PIK3R2 | 7997 | 0.358 | -0.3393 | No | ||
| 10 | PLCE1 | 8351 | 0.267 | -0.3567 | No | ||
| 11 | STAT5A | 8412 | 0.253 | -0.3583 | No | ||
| 12 | KIT | 8853 | 0.149 | -0.3811 | No | ||
| 13 | HRAS | 9216 | 0.053 | -0.4003 | No | ||
| 14 | PLCG1 | 9738 | -0.081 | -0.4278 | No | ||
| 15 | LYN | 9815 | -0.105 | -0.4313 | No | ||
| 16 | STAT5B | 9990 | -0.149 | -0.4397 | No | ||
| 17 | SOS1 | 10324 | -0.226 | -0.4562 | No | ||
| 18 | CBL | 10381 | -0.236 | -0.4578 | No | ||
| 19 | GRAP | 10568 | -0.278 | -0.4661 | No | ||
| 20 | MITF | 12395 | -0.766 | -0.5596 | No | ||
| 21 | RASA1 | 12828 | -0.895 | -0.5773 | No | ||
| 22 | PTPN11 | 13063 | -0.961 | -0.5840 | No | ||
| 23 | SPRED1 | 13275 | -1.041 | -0.5889 | No | ||
| 24 | SOCS6 | 14007 | -1.323 | -0.6200 | No | ||
| 25 | YES1 | 14205 | -1.402 | -0.6219 | No | ||
| 26 | EPOR | 14555 | -1.547 | -0.6311 | No | ||
| 27 | KITLG | 14610 | -1.576 | -0.6242 | No | ||
| 28 | RAF1 | 14806 | -1.685 | -0.6242 | No | ||
| 29 | SHC1 | 14889 | -1.736 | -0.6178 | No | ||
| 30 | ABL1 | 15100 | -1.861 | -0.6175 | No | ||
| 31 | FYN | 16296 | -3.029 | -0.6630 | Yes | ||
| 32 | GRB2 | 16640 | -3.636 | -0.6589 | Yes | ||
| 33 | JAK2 | 16656 | -3.667 | -0.6369 | Yes | ||
| 34 | FGR | 16670 | -3.687 | -0.6146 | Yes | ||
| 35 | MATK | 16846 | -4.009 | -0.5991 | Yes | ||
| 36 | BAD | 17107 | -4.710 | -0.5838 | Yes | ||
| 37 | CRKL | 17222 | -5.001 | -0.5589 | Yes | ||
| 38 | FES | 17409 | -5.547 | -0.5344 | Yes | ||
| 39 | HCK | 17570 | -6.125 | -0.5049 | Yes | ||
| 40 | SH3KBP1 | 17801 | -6.943 | -0.4741 | Yes | ||
| 41 | DOK1 | 17867 | -7.248 | -0.4325 | Yes | ||
| 42 | CBLB | 17978 | -7.787 | -0.3900 | Yes | ||
| 43 | VAV1 | 18125 | -8.843 | -0.3428 | Yes | ||
| 44 | SOCS5 | 18141 | -9.002 | -0.2876 | Yes | ||
| 45 | PIK3CG | 18147 | -9.020 | -0.2318 | Yes | ||
| 46 | PRKCA | 18153 | -9.054 | -0.1757 | Yes | ||
| 47 | SOCS1 | 18162 | -9.161 | -0.1191 | Yes | ||
| 48 | STAT1 | 18246 | -9.978 | -0.0615 | Yes | ||
| 49 | PTPN6 | 18441 | -13.086 | 0.0094 | Yes |