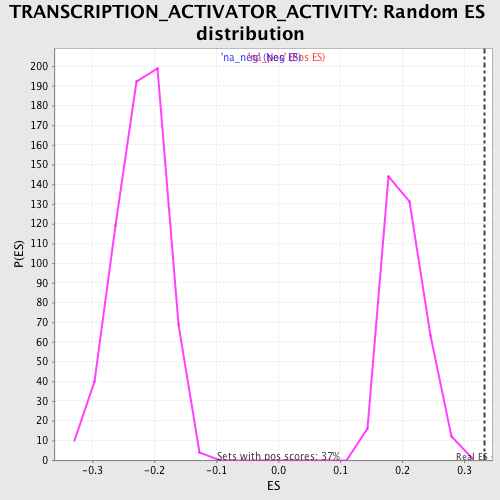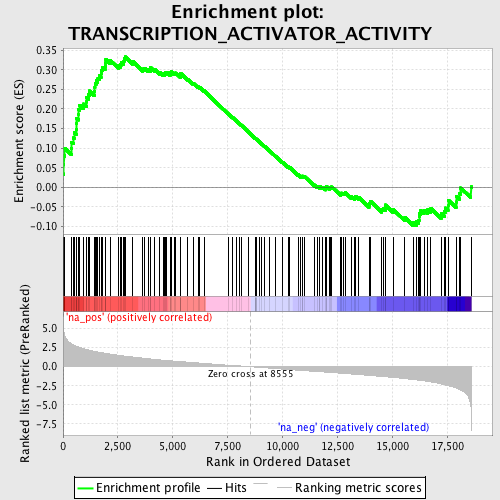
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TRANSCRIPTION_ACTIVATOR_ACTIVITY |
| Enrichment Score (ES) | 0.3329411 |
| Normalized Enrichment Score (NES) | 1.6328834 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.48612309 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TCERG1 | 0 | 6.442 | 0.0343 | Yes | ||
| 2 | PPARGC1B | 27 | 4.654 | 0.0577 | Yes | ||
| 3 | HCFC1 | 40 | 4.444 | 0.0807 | Yes | ||
| 4 | E4F1 | 76 | 3.988 | 0.1001 | Yes | ||
| 5 | ERCC2 | 372 | 3.000 | 0.1001 | Yes | ||
| 6 | ARID1A | 388 | 2.976 | 0.1152 | Yes | ||
| 7 | NFATC1 | 467 | 2.872 | 0.1262 | Yes | ||
| 8 | SMARCD3 | 505 | 2.821 | 0.1393 | Yes | ||
| 9 | GTF2F1 | 598 | 2.697 | 0.1487 | Yes | ||
| 10 | EPC1 | 609 | 2.691 | 0.1625 | Yes | ||
| 11 | BRD8 | 618 | 2.683 | 0.1763 | Yes | ||
| 12 | NFKB2 | 704 | 2.580 | 0.1855 | Yes | ||
| 13 | CIITA | 712 | 2.570 | 0.1988 | Yes | ||
| 14 | ATF4 | 752 | 2.518 | 0.2101 | Yes | ||
| 15 | GCN5L2 | 936 | 2.361 | 0.2128 | Yes | ||
| 16 | CREBBP | 1062 | 2.275 | 0.2181 | Yes | ||
| 17 | MAX | 1072 | 2.267 | 0.2297 | Yes | ||
| 18 | CXXC1 | 1164 | 2.182 | 0.2364 | Yes | ||
| 19 | MYST4 | 1199 | 2.157 | 0.2461 | Yes | ||
| 20 | ELF4 | 1419 | 2.012 | 0.2450 | Yes | ||
| 21 | RSF1 | 1446 | 1.998 | 0.2542 | Yes | ||
| 22 | POU2AF1 | 1454 | 1.996 | 0.2645 | Yes | ||
| 23 | YY1 | 1503 | 1.970 | 0.2724 | Yes | ||
| 24 | RXRB | 1586 | 1.919 | 0.2781 | Yes | ||
| 25 | SMARCA4 | 1648 | 1.884 | 0.2849 | Yes | ||
| 26 | ILF3 | 1731 | 1.839 | 0.2902 | Yes | ||
| 27 | NCOA6 | 1746 | 1.829 | 0.2992 | Yes | ||
| 28 | MYOD1 | 1788 | 1.816 | 0.3067 | Yes | ||
| 29 | ATF2 | 1915 | 1.751 | 0.3092 | Yes | ||
| 30 | MAML1 | 1935 | 1.740 | 0.3175 | Yes | ||
| 31 | CARM1 | 1946 | 1.732 | 0.3261 | Yes | ||
| 32 | MTF1 | 2156 | 1.647 | 0.3236 | Yes | ||
| 33 | GLIS2 | 2521 | 1.499 | 0.3119 | Yes | ||
| 34 | SMARCD1 | 2619 | 1.457 | 0.3144 | Yes | ||
| 35 | RFXAP | 2665 | 1.443 | 0.3197 | Yes | ||
| 36 | BRCA2 | 2768 | 1.402 | 0.3216 | Yes | ||
| 37 | PAX8 | 2780 | 1.397 | 0.3285 | Yes | ||
| 38 | TAF4 | 2834 | 1.377 | 0.3329 | Yes | ||
| 39 | UTF1 | 3184 | 1.262 | 0.3208 | No | ||
| 40 | ARNT | 3640 | 1.113 | 0.3021 | No | ||
| 41 | NFE2 | 3708 | 1.087 | 0.3043 | No | ||
| 42 | RUNX2 | 3878 | 1.034 | 0.3006 | No | ||
| 43 | MED6 | 3977 | 1.008 | 0.3007 | No | ||
| 44 | TGFB1I1 | 3981 | 1.008 | 0.3059 | No | ||
| 45 | NFATC2 | 4161 | 0.959 | 0.3013 | No | ||
| 46 | UBE2V1 | 4399 | 0.899 | 0.2933 | No | ||
| 47 | GLI1 | 4559 | 0.854 | 0.2893 | No | ||
| 48 | ELF3 | 4624 | 0.841 | 0.2903 | No | ||
| 49 | PHB | 4668 | 0.831 | 0.2924 | No | ||
| 50 | MYEF2 | 4718 | 0.817 | 0.2941 | No | ||
| 51 | SMARCD2 | 4877 | 0.775 | 0.2897 | No | ||
| 52 | NR1H3 | 4878 | 0.775 | 0.2938 | No | ||
| 53 | KLF6 | 4926 | 0.765 | 0.2953 | No | ||
| 54 | NFKBIB | 5066 | 0.732 | 0.2917 | No | ||
| 55 | RXRA | 5134 | 0.716 | 0.2919 | No | ||
| 56 | NR1I3 | 5350 | 0.662 | 0.2838 | No | ||
| 57 | SP1 | 5357 | 0.661 | 0.2870 | No | ||
| 58 | SOX10 | 5361 | 0.660 | 0.2903 | No | ||
| 59 | NFYC | 5678 | 0.596 | 0.2764 | No | ||
| 60 | SMARCE1 | 5932 | 0.541 | 0.2656 | No | ||
| 61 | HMGA1 | 6149 | 0.492 | 0.2565 | No | ||
| 62 | KLF7 | 6236 | 0.473 | 0.2544 | No | ||
| 63 | RARA | 6426 | 0.429 | 0.2465 | No | ||
| 64 | ZEB1 | 7542 | 0.193 | 0.1872 | No | ||
| 65 | TBPL1 | 7733 | 0.160 | 0.1777 | No | ||
| 66 | TFEC | 7883 | 0.130 | 0.1704 | No | ||
| 67 | NR2C2 | 8036 | 0.103 | 0.1627 | No | ||
| 68 | EPAS1 | 8111 | 0.089 | 0.1592 | No | ||
| 69 | CDH1 | 8427 | 0.026 | 0.1423 | No | ||
| 70 | FUBP3 | 8747 | -0.040 | 0.1252 | No | ||
| 71 | ZNF148 | 8778 | -0.045 | 0.1238 | No | ||
| 72 | GMEB1 | 8786 | -0.047 | 0.1237 | No | ||
| 73 | MNT | 8801 | -0.050 | 0.1232 | No | ||
| 74 | TGFB1 | 8803 | -0.050 | 0.1234 | No | ||
| 75 | NPM1 | 8957 | -0.083 | 0.1156 | No | ||
| 76 | ESRRG | 9060 | -0.103 | 0.1106 | No | ||
| 77 | MITF | 9171 | -0.128 | 0.1054 | No | ||
| 78 | HIVEP3 | 9404 | -0.178 | 0.0938 | No | ||
| 79 | SP4 | 9686 | -0.238 | 0.0798 | No | ||
| 80 | PMF1 | 10000 | -0.308 | 0.0645 | No | ||
| 81 | PPARGC1A | 10259 | -0.366 | 0.0525 | No | ||
| 82 | FOXC2 | 10325 | -0.380 | 0.0510 | No | ||
| 83 | NFE2L3 | 10710 | -0.454 | 0.0327 | No | ||
| 84 | EDF1 | 10840 | -0.482 | 0.0283 | No | ||
| 85 | MKL2 | 10891 | -0.494 | 0.0282 | No | ||
| 86 | NRIP1 | 10925 | -0.501 | 0.0291 | No | ||
| 87 | NFATC4 | 11005 | -0.520 | 0.0276 | No | ||
| 88 | JMY | 11460 | -0.607 | 0.0062 | No | ||
| 89 | PQBP1 | 11582 | -0.628 | 0.0030 | No | ||
| 90 | JUNB | 11679 | -0.647 | 0.0013 | No | ||
| 91 | YWHAH | 11704 | -0.651 | 0.0035 | No | ||
| 92 | SMAD2 | 11833 | -0.678 | 0.0001 | No | ||
| 93 | SMARCC1 | 11960 | -0.701 | -0.0029 | No | ||
| 94 | PCBD1 | 11983 | -0.705 | -0.0004 | No | ||
| 95 | TFDP1 | 11988 | -0.705 | 0.0032 | No | ||
| 96 | GABPA | 12156 | -0.737 | -0.0019 | No | ||
| 97 | BRDT | 12183 | -0.743 | 0.0006 | No | ||
| 98 | AIP | 12247 | -0.759 | 0.0012 | No | ||
| 99 | FOXO3 | 12647 | -0.847 | -0.0158 | No | ||
| 100 | RNF14 | 12704 | -0.860 | -0.0143 | No | ||
| 101 | TRIM24 | 12799 | -0.879 | -0.0147 | No | ||
| 102 | GLI2 | 12851 | -0.889 | -0.0127 | No | ||
| 103 | NR1H4 | 13136 | -0.949 | -0.0230 | No | ||
| 104 | TRIP4 | 13290 | -0.983 | -0.0260 | No | ||
| 105 | VGLL1 | 13335 | -0.995 | -0.0231 | No | ||
| 106 | RUNX1 | 13479 | -1.027 | -0.0254 | No | ||
| 107 | NR1I2 | 13943 | -1.135 | -0.0444 | No | ||
| 108 | SNW1 | 13973 | -1.142 | -0.0399 | No | ||
| 109 | FOXH1 | 14008 | -1.150 | -0.0356 | No | ||
| 110 | PRRX1 | 14518 | -1.273 | -0.0564 | No | ||
| 111 | ELF2 | 14580 | -1.287 | -0.0528 | No | ||
| 112 | PSMC3 | 14692 | -1.312 | -0.0518 | No | ||
| 113 | APEX1 | 14693 | -1.313 | -0.0448 | No | ||
| 114 | DYRK1B | 15059 | -1.401 | -0.0571 | No | ||
| 115 | ESR2 | 15579 | -1.556 | -0.0769 | No | ||
| 116 | PCAF | 15978 | -1.689 | -0.0894 | No | ||
| 117 | BCL10 | 16112 | -1.737 | -0.0874 | No | ||
| 118 | ABT1 | 16212 | -1.769 | -0.0833 | No | ||
| 119 | TAF9 | 16227 | -1.773 | -0.0746 | No | ||
| 120 | TRIM32 | 16244 | -1.778 | -0.0660 | No | ||
| 121 | DTX1 | 16302 | -1.800 | -0.0595 | No | ||
| 122 | GTF2A1 | 16452 | -1.850 | -0.0577 | No | ||
| 123 | HTATIP2 | 16624 | -1.928 | -0.0567 | No | ||
| 124 | NR2F1 | 16761 | -1.987 | -0.0534 | No | ||
| 125 | RBM14 | 17230 | -2.247 | -0.0668 | No | ||
| 126 | ZFX | 17376 | -2.347 | -0.0621 | No | ||
| 127 | YAF2 | 17434 | -2.388 | -0.0525 | No | ||
| 128 | NR5A1 | 17546 | -2.454 | -0.0454 | No | ||
| 129 | HSF2 | 17555 | -2.459 | -0.0327 | No | ||
| 130 | MED4 | 17911 | -2.780 | -0.0371 | No | ||
| 131 | COPS5 | 17919 | -2.789 | -0.0227 | No | ||
| 132 | TAF7 | 18064 | -2.978 | -0.0146 | No | ||
| 133 | RIPK3 | 18124 | -3.066 | -0.0014 | No | ||
| 134 | IRF4 | 18590 | -5.255 | 0.0014 | No |