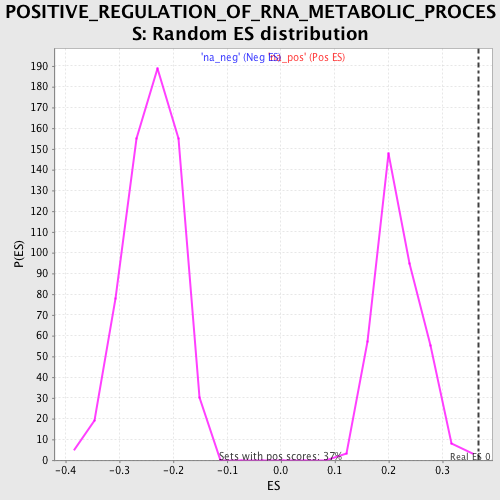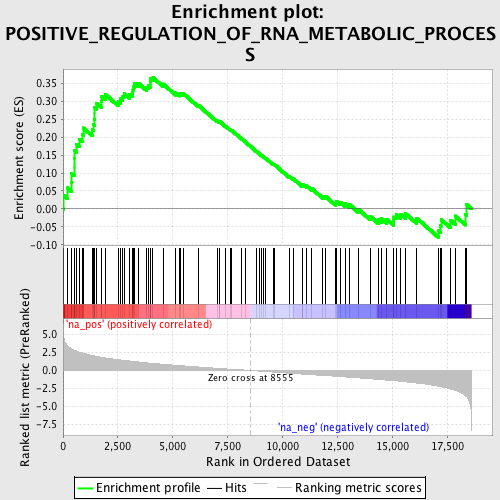
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | POSITIVE_REGULATION_OF_RNA_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.36650643 |
| Normalized Enrichment Score (NES) | 1.6720364 |
| Nominal p-value | 0.0027100272 |
| FDR q-value | 0.49418452 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PPARGC1B | 27 | 4.654 | 0.0378 | Yes | ||
| 2 | NSD1 | 186 | 3.492 | 0.0587 | Yes | ||
| 3 | ERCC2 | 372 | 3.000 | 0.0740 | Yes | ||
| 4 | ARID1A | 388 | 2.976 | 0.0983 | Yes | ||
| 5 | ATF7IP | 500 | 2.826 | 0.1161 | Yes | ||
| 6 | TP53 | 501 | 2.825 | 0.1400 | Yes | ||
| 7 | SMARCD3 | 505 | 2.821 | 0.1636 | Yes | ||
| 8 | EPC1 | 609 | 2.691 | 0.1807 | Yes | ||
| 9 | ATF4 | 752 | 2.518 | 0.1943 | Yes | ||
| 10 | ARHGEF11 | 865 | 2.427 | 0.2087 | Yes | ||
| 11 | CTCF | 918 | 2.376 | 0.2259 | Yes | ||
| 12 | MED12 | 1326 | 2.063 | 0.2214 | Yes | ||
| 13 | CLOCK | 1378 | 2.031 | 0.2357 | Yes | ||
| 14 | ELF4 | 1419 | 2.012 | 0.2505 | Yes | ||
| 15 | TCF4 | 1445 | 1.998 | 0.2660 | Yes | ||
| 16 | RSF1 | 1446 | 1.998 | 0.2829 | Yes | ||
| 17 | GTF2H1 | 1526 | 1.955 | 0.2951 | Yes | ||
| 18 | ILF3 | 1731 | 1.839 | 0.2996 | Yes | ||
| 19 | NCOA6 | 1746 | 1.829 | 0.3143 | Yes | ||
| 20 | MAML1 | 1935 | 1.740 | 0.3188 | Yes | ||
| 21 | BPTF | 2508 | 1.503 | 0.3006 | Yes | ||
| 22 | SMARCD1 | 2619 | 1.457 | 0.3069 | Yes | ||
| 23 | EHF | 2723 | 1.420 | 0.3134 | Yes | ||
| 24 | PAX8 | 2780 | 1.397 | 0.3221 | Yes | ||
| 25 | TNFRSF1A | 3037 | 1.309 | 0.3193 | Yes | ||
| 26 | NOTCH4 | 3144 | 1.276 | 0.3244 | Yes | ||
| 27 | UTF1 | 3184 | 1.262 | 0.3329 | Yes | ||
| 28 | HNF4A | 3225 | 1.247 | 0.3413 | Yes | ||
| 29 | ELL3 | 3252 | 1.235 | 0.3503 | Yes | ||
| 30 | FOXF2 | 3434 | 1.179 | 0.3504 | Yes | ||
| 31 | IL4 | 3814 | 1.054 | 0.3389 | Yes | ||
| 32 | MAP2K3 | 3890 | 1.029 | 0.3435 | Yes | ||
| 33 | SUPT5H | 3964 | 1.011 | 0.3481 | Yes | ||
| 34 | NARG1 | 3966 | 1.010 | 0.3566 | Yes | ||
| 35 | MED6 | 3977 | 1.008 | 0.3645 | Yes | ||
| 36 | GLIS1 | 4094 | 0.978 | 0.3665 | Yes | ||
| 37 | GLI1 | 4559 | 0.854 | 0.3487 | No | ||
| 38 | RXRA | 5134 | 0.716 | 0.3237 | No | ||
| 39 | SMAD3 | 5295 | 0.677 | 0.3208 | No | ||
| 40 | SP1 | 5357 | 0.661 | 0.3231 | No | ||
| 41 | ELF1 | 5490 | 0.632 | 0.3213 | No | ||
| 42 | HMGA1 | 6149 | 0.492 | 0.2899 | No | ||
| 43 | HIF1A | 7018 | 0.299 | 0.2456 | No | ||
| 44 | MNAT1 | 7120 | 0.278 | 0.2425 | No | ||
| 45 | FOXE1 | 7131 | 0.275 | 0.2443 | No | ||
| 46 | PHF5A | 7423 | 0.215 | 0.2304 | No | ||
| 47 | THRAP3 | 7647 | 0.175 | 0.2198 | No | ||
| 48 | RORB | 7671 | 0.172 | 0.2200 | No | ||
| 49 | EPAS1 | 8111 | 0.089 | 0.1971 | No | ||
| 50 | HNRPAB | 8309 | 0.054 | 0.1869 | No | ||
| 51 | TGFB1 | 8803 | -0.050 | 0.1607 | No | ||
| 52 | ARNTL | 8961 | -0.084 | 0.1530 | No | ||
| 53 | ESRRG | 9060 | -0.103 | 0.1485 | No | ||
| 54 | TCF3 | 9110 | -0.115 | 0.1469 | No | ||
| 55 | SCAP | 9245 | -0.147 | 0.1409 | No | ||
| 56 | CDK7 | 9570 | -0.214 | 0.1252 | No | ||
| 57 | SPI1 | 9616 | -0.224 | 0.1246 | No | ||
| 58 | PLAGL1 | 10314 | -0.378 | 0.0902 | No | ||
| 59 | NPAS2 | 10490 | -0.410 | 0.0842 | No | ||
| 60 | MKL2 | 10891 | -0.494 | 0.0668 | No | ||
| 61 | NRIP1 | 10925 | -0.501 | 0.0692 | No | ||
| 62 | TBX5 | 11088 | -0.535 | 0.0650 | No | ||
| 63 | BMP6 | 11342 | -0.583 | 0.0563 | No | ||
| 64 | SMAD2 | 11833 | -0.678 | 0.0355 | No | ||
| 65 | SMARCC1 | 11960 | -0.701 | 0.0346 | No | ||
| 66 | FOXD3 | 12420 | -0.794 | 0.0166 | No | ||
| 67 | GATA4 | 12454 | -0.801 | 0.0215 | No | ||
| 68 | FOXO3 | 12647 | -0.847 | 0.0183 | No | ||
| 69 | GLI2 | 12851 | -0.889 | 0.0149 | No | ||
| 70 | TRIM28 | 13051 | -0.932 | 0.0120 | No | ||
| 71 | RUNX1 | 13479 | -1.027 | -0.0024 | No | ||
| 72 | FOXH1 | 14008 | -1.150 | -0.0212 | No | ||
| 73 | MYO6 | 14365 | -1.236 | -0.0300 | No | ||
| 74 | TRERF1 | 14516 | -1.273 | -0.0274 | No | ||
| 75 | NUFIP1 | 14758 | -1.327 | -0.0292 | No | ||
| 76 | DYRK1B | 15059 | -1.401 | -0.0336 | No | ||
| 77 | MRPL12 | 15076 | -1.406 | -0.0226 | No | ||
| 78 | CCNH | 15175 | -1.431 | -0.0158 | No | ||
| 79 | SMARCA1 | 15386 | -1.496 | -0.0145 | No | ||
| 80 | CITED2 | 15609 | -1.567 | -0.0133 | No | ||
| 81 | BCL10 | 16112 | -1.737 | -0.0257 | No | ||
| 82 | XRCC6 | 17113 | -2.175 | -0.0613 | No | ||
| 83 | CREB5 | 17187 | -2.220 | -0.0466 | No | ||
| 84 | RBM14 | 17230 | -2.247 | -0.0299 | No | ||
| 85 | ERCC3 | 17651 | -2.555 | -0.0310 | No | ||
| 86 | SQSTM1 | 17871 | -2.744 | -0.0197 | No | ||
| 87 | ILF2 | 18341 | -3.481 | -0.0157 | No | ||
| 88 | SUPT4H1 | 18386 | -3.610 | 0.0124 | No |