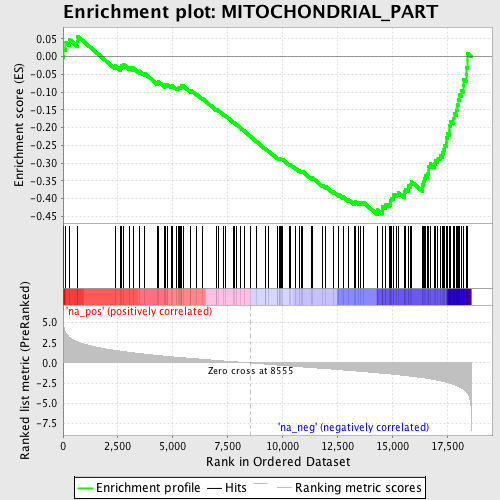
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | MITOCHONDRIAL_PART |
| Enrichment Score (ES) | -0.44462183 |
| Normalized Enrichment Score (NES) | -1.9842393 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.025153179 |
| FWER p-Value | 0.153 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BCL2 | 39 | 4.461 | 0.0230 | No | ||
| 2 | SLC25A22 | 129 | 3.696 | 0.0389 | No | ||
| 3 | NFS1 | 286 | 3.177 | 0.0483 | No | ||
| 4 | OPA1 | 650 | 2.631 | 0.0435 | No | ||
| 5 | MCL1 | 659 | 2.620 | 0.0578 | No | ||
| 6 | PIN4 | 2370 | 1.556 | -0.0260 | No | ||
| 7 | RHOT2 | 2603 | 1.465 | -0.0303 | No | ||
| 8 | PPOX | 2673 | 1.439 | -0.0259 | No | ||
| 9 | RAF1 | 2767 | 1.403 | -0.0231 | No | ||
| 10 | IMMT | 3020 | 1.314 | -0.0293 | No | ||
| 11 | TIMM10 | 3196 | 1.254 | -0.0318 | No | ||
| 12 | HTRA2 | 3478 | 1.166 | -0.0404 | No | ||
| 13 | CENTA2 | 3711 | 1.087 | -0.0468 | No | ||
| 14 | MFN2 | 4302 | 0.925 | -0.0736 | No | ||
| 15 | MRPS22 | 4324 | 0.920 | -0.0695 | No | ||
| 16 | HCCS | 4626 | 0.840 | -0.0811 | No | ||
| 17 | PHB | 4668 | 0.831 | -0.0786 | No | ||
| 18 | POLG2 | 4761 | 0.805 | -0.0791 | No | ||
| 19 | NDUFS1 | 4920 | 0.766 | -0.0833 | No | ||
| 20 | NR3C1 | 4965 | 0.755 | -0.0815 | No | ||
| 21 | MTX2 | 5171 | 0.708 | -0.0886 | No | ||
| 22 | CASP7 | 5238 | 0.693 | -0.0882 | No | ||
| 23 | MRPS15 | 5284 | 0.680 | -0.0868 | No | ||
| 24 | MRPS12 | 5341 | 0.665 | -0.0861 | No | ||
| 25 | TIMM44 | 5379 | 0.655 | -0.0845 | No | ||
| 26 | ATP5B | 5397 | 0.651 | -0.0817 | No | ||
| 27 | SLC25A3 | 5465 | 0.637 | -0.0818 | No | ||
| 28 | ALDH4A1 | 5814 | 0.568 | -0.0974 | No | ||
| 29 | TOMM34 | 5825 | 0.565 | -0.0948 | No | ||
| 30 | ATP5A1 | 6077 | 0.508 | -0.1055 | No | ||
| 31 | TIMM23 | 6368 | 0.442 | -0.1187 | No | ||
| 32 | COX6B2 | 6971 | 0.309 | -0.1495 | No | ||
| 33 | ATP5D | 6997 | 0.304 | -0.1492 | No | ||
| 34 | HSD3B1 | 7099 | 0.282 | -0.1530 | No | ||
| 35 | MRPL23 | 7324 | 0.237 | -0.1638 | No | ||
| 36 | GATM | 7422 | 0.215 | -0.1679 | No | ||
| 37 | SDHA | 7744 | 0.158 | -0.1843 | No | ||
| 38 | BNIP3 | 7791 | 0.149 | -0.1860 | No | ||
| 39 | ALAS2 | 7918 | 0.123 | -0.1921 | No | ||
| 40 | UQCRB | 8089 | 0.092 | -0.2008 | No | ||
| 41 | MRPS24 | 8264 | 0.062 | -0.2098 | No | ||
| 42 | UQCRC1 | 8522 | 0.006 | -0.2237 | No | ||
| 43 | SDHD | 8824 | -0.054 | -0.2397 | No | ||
| 44 | NDUFA6 | 9214 | -0.139 | -0.2600 | No | ||
| 45 | NDUFS7 | 9216 | -0.140 | -0.2592 | No | ||
| 46 | RAB11FIP5 | 9380 | -0.174 | -0.2671 | No | ||
| 47 | MRPL52 | 9759 | -0.257 | -0.2861 | No | ||
| 48 | MRPL51 | 9782 | -0.262 | -0.2858 | No | ||
| 49 | NDUFA13 | 9842 | -0.272 | -0.2874 | No | ||
| 50 | ATP5C1 | 9856 | -0.275 | -0.2866 | No | ||
| 51 | MRPL32 | 9925 | -0.290 | -0.2886 | No | ||
| 52 | BCKDHB | 9958 | -0.298 | -0.2887 | No | ||
| 53 | CYCS | 9987 | -0.305 | -0.2885 | No | ||
| 54 | TIMM17B | 10329 | -0.381 | -0.3048 | No | ||
| 55 | CS | 10365 | -0.387 | -0.3045 | No | ||
| 56 | OGDH | 10608 | -0.434 | -0.3152 | No | ||
| 57 | TIMM9 | 10764 | -0.467 | -0.3209 | No | ||
| 58 | TFB2M | 10862 | -0.486 | -0.3234 | No | ||
| 59 | ETFB | 10909 | -0.497 | -0.3231 | No | ||
| 60 | VDAC1 | 11327 | -0.580 | -0.3424 | No | ||
| 61 | ACADM | 11357 | -0.585 | -0.3407 | No | ||
| 62 | ATP5O | 11826 | -0.676 | -0.3622 | No | ||
| 63 | ATP5G3 | 11977 | -0.704 | -0.3664 | No | ||
| 64 | ATP5G2 | 12328 | -0.777 | -0.3810 | No | ||
| 65 | NDUFAB1 | 12549 | -0.824 | -0.3882 | No | ||
| 66 | SUPV3L1 | 12756 | -0.871 | -0.3945 | No | ||
| 67 | ATP5E | 12996 | -0.921 | -0.4022 | No | ||
| 68 | ABCF2 | 13282 | -0.982 | -0.4121 | No | ||
| 69 | MRPS18C | 13316 | -0.990 | -0.4083 | No | ||
| 70 | MRPS36 | 13449 | -1.019 | -0.4097 | No | ||
| 71 | SLC25A11 | 13565 | -1.045 | -0.4101 | No | ||
| 72 | DBT | 13697 | -1.075 | -0.4111 | No | ||
| 73 | PHB2 | 14317 | -1.224 | -0.4377 | Yes | ||
| 74 | NDUFS8 | 14334 | -1.229 | -0.4317 | Yes | ||
| 75 | ATP5G1 | 14549 | -1.282 | -0.4361 | Yes | ||
| 76 | SLC25A1 | 14554 | -1.282 | -0.4291 | Yes | ||
| 77 | MAOB | 14560 | -1.283 | -0.4221 | Yes | ||
| 78 | MRPS16 | 14675 | -1.308 | -0.4210 | Yes | ||
| 79 | VDAC3 | 14712 | -1.317 | -0.4155 | Yes | ||
| 80 | ATP5F1 | 14860 | -1.351 | -0.4159 | Yes | ||
| 81 | UQCRH | 14927 | -1.368 | -0.4117 | Yes | ||
| 82 | UCP3 | 14935 | -1.371 | -0.4044 | Yes | ||
| 83 | MRPS28 | 14969 | -1.384 | -0.3984 | Yes | ||
| 84 | RHOT1 | 15040 | -1.397 | -0.3944 | Yes | ||
| 85 | MRPL12 | 15076 | -1.406 | -0.3884 | Yes | ||
| 86 | VDAC2 | 15201 | -1.438 | -0.3870 | Yes | ||
| 87 | TOMM22 | 15270 | -1.456 | -0.3825 | Yes | ||
| 88 | MRPS18A | 15546 | -1.546 | -0.3887 | Yes | ||
| 89 | NDUFA1 | 15551 | -1.547 | -0.3802 | Yes | ||
| 90 | COX15 | 15601 | -1.563 | -0.3741 | Yes | ||
| 91 | PMPCA | 15729 | -1.603 | -0.3719 | Yes | ||
| 92 | MRPS21 | 15746 | -1.609 | -0.3637 | Yes | ||
| 93 | DNAJA3 | 15828 | -1.635 | -0.3589 | Yes | ||
| 94 | BCKDK | 15868 | -1.650 | -0.3518 | Yes | ||
| 95 | MRPL55 | 16377 | -1.825 | -0.3690 | Yes | ||
| 96 | MRPL10 | 16382 | -1.826 | -0.3589 | Yes | ||
| 97 | FIS1 | 16443 | -1.848 | -0.3518 | Yes | ||
| 98 | PITRM1 | 16465 | -1.856 | -0.3425 | Yes | ||
| 99 | NDUFV1 | 16516 | -1.880 | -0.3347 | Yes | ||
| 100 | AIFM2 | 16627 | -1.929 | -0.3298 | Yes | ||
| 101 | TIMM8B | 16656 | -1.944 | -0.3204 | Yes | ||
| 102 | ETFA | 16658 | -1.944 | -0.3095 | Yes | ||
| 103 | MRPL40 | 16725 | -1.973 | -0.3020 | Yes | ||
| 104 | NDUFS3 | 16932 | -2.073 | -0.3015 | Yes | ||
| 105 | TIMM17A | 16967 | -2.090 | -0.2916 | Yes | ||
| 106 | NDUFS2 | 17085 | -2.153 | -0.2858 | Yes | ||
| 107 | CASQ1 | 17185 | -2.218 | -0.2787 | Yes | ||
| 108 | NDUFA2 | 17274 | -2.274 | -0.2707 | Yes | ||
| 109 | GRPEL1 | 17356 | -2.335 | -0.2619 | Yes | ||
| 110 | HSD3B2 | 17385 | -2.354 | -0.2502 | Yes | ||
| 111 | ABCB7 | 17462 | -2.405 | -0.2408 | Yes | ||
| 112 | MPV17 | 17467 | -2.406 | -0.2275 | Yes | ||
| 113 | TIMM50 | 17527 | -2.438 | -0.2170 | Yes | ||
| 114 | ATP5J | 17623 | -2.531 | -0.2079 | Yes | ||
| 115 | NDUFS4 | 17625 | -2.531 | -0.1937 | Yes | ||
| 116 | BCS1L | 17664 | -2.564 | -0.1814 | Yes | ||
| 117 | NNT | 17791 | -2.673 | -0.1732 | Yes | ||
| 118 | TIMM13 | 17817 | -2.690 | -0.1594 | Yes | ||
| 119 | HADHB | 17943 | -2.825 | -0.1503 | Yes | ||
| 120 | NDUFA9 | 17956 | -2.841 | -0.1350 | Yes | ||
| 121 | BAX | 18002 | -2.899 | -0.1211 | Yes | ||
| 122 | SURF1 | 18068 | -2.986 | -0.1078 | Yes | ||
| 123 | BCKDHA | 18168 | -3.133 | -0.0956 | Yes | ||
| 124 | ACN9 | 18246 | -3.256 | -0.0814 | Yes | ||
| 125 | MRPS35 | 18262 | -3.290 | -0.0638 | Yes | ||
| 126 | MRPS10 | 18366 | -3.568 | -0.0493 | Yes | ||
| 127 | SLC25A15 | 18388 | -3.611 | -0.0301 | Yes | ||
| 128 | MRPS11 | 18437 | -3.763 | -0.0116 | Yes | ||
| 129 | ABCB6 | 18438 | -3.774 | 0.0096 | Yes |