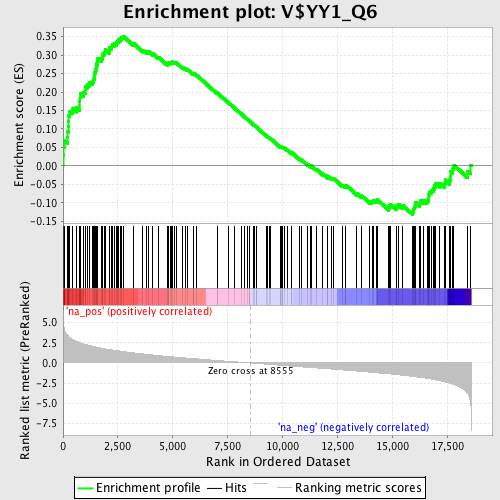
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$YY1_Q6 |
| Enrichment Score (ES) | 0.3515126 |
| Normalized Enrichment Score (NES) | 1.7501631 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07508443 |
| FWER p-Value | 0.295 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CCND1 | 2 | 6.319 | 0.0296 | Yes | ||
| 2 | TRIM8 | 19 | 4.785 | 0.0512 | Yes | ||
| 3 | BCL11A | 59 | 4.158 | 0.0686 | Yes | ||
| 4 | UBE3A | 193 | 3.467 | 0.0777 | Yes | ||
| 5 | RBM3 | 212 | 3.392 | 0.0926 | Yes | ||
| 6 | AP3D1 | 235 | 3.306 | 0.1070 | Yes | ||
| 7 | TNRC15 | 251 | 3.257 | 0.1215 | Yes | ||
| 8 | USF1 | 258 | 3.239 | 0.1363 | Yes | ||
| 9 | ASH1L | 311 | 3.130 | 0.1482 | Yes | ||
| 10 | IRAK1 | 447 | 2.898 | 0.1545 | Yes | ||
| 11 | EPC1 | 609 | 2.691 | 0.1585 | Yes | ||
| 12 | ATF4 | 752 | 2.518 | 0.1626 | Yes | ||
| 13 | NCOR1 | 754 | 2.513 | 0.1743 | Yes | ||
| 14 | ABHD1 | 769 | 2.502 | 0.1853 | Yes | ||
| 15 | UBE1 | 784 | 2.491 | 0.1963 | Yes | ||
| 16 | LPHN1 | 928 | 2.368 | 0.1997 | Yes | ||
| 17 | CLK2 | 1018 | 2.306 | 0.2057 | Yes | ||
| 18 | DDX6 | 1021 | 2.305 | 0.2164 | Yes | ||
| 19 | GTPBP1 | 1129 | 2.208 | 0.2210 | Yes | ||
| 20 | HSF1 | 1218 | 2.141 | 0.2263 | Yes | ||
| 21 | POU2F1 | 1339 | 2.053 | 0.2294 | Yes | ||
| 22 | GIT2 | 1403 | 2.020 | 0.2355 | Yes | ||
| 23 | SF3A1 | 1425 | 2.009 | 0.2438 | Yes | ||
| 24 | TCF4 | 1445 | 1.998 | 0.2521 | Yes | ||
| 25 | CASC3 | 1466 | 1.989 | 0.2604 | Yes | ||
| 26 | YY1 | 1503 | 1.970 | 0.2677 | Yes | ||
| 27 | UBE4B | 1511 | 1.963 | 0.2765 | Yes | ||
| 28 | RAB22A | 1580 | 1.921 | 0.2819 | Yes | ||
| 29 | RXRB | 1586 | 1.919 | 0.2906 | Yes | ||
| 30 | SNRPN | 1741 | 1.833 | 0.2909 | Yes | ||
| 31 | CNOT3 | 1793 | 1.812 | 0.2966 | Yes | ||
| 32 | HIC2 | 1816 | 1.801 | 0.3039 | Yes | ||
| 33 | PTBP1 | 1890 | 1.765 | 0.3082 | Yes | ||
| 34 | PRKCSH | 1912 | 1.753 | 0.3153 | Yes | ||
| 35 | PFN2 | 2097 | 1.676 | 0.3132 | Yes | ||
| 36 | PUM1 | 2103 | 1.671 | 0.3208 | Yes | ||
| 37 | A2BP1 | 2193 | 1.632 | 0.3237 | Yes | ||
| 38 | DGCR2 | 2228 | 1.615 | 0.3294 | Yes | ||
| 39 | NASP | 2327 | 1.573 | 0.3315 | Yes | ||
| 40 | SFPQ | 2424 | 1.532 | 0.3335 | Yes | ||
| 41 | SNX5 | 2484 | 1.509 | 0.3374 | Yes | ||
| 42 | TGM5 | 2528 | 1.497 | 0.3421 | Yes | ||
| 43 | HMGB3 | 2616 | 1.460 | 0.3442 | Yes | ||
| 44 | DSCAM | 2654 | 1.445 | 0.3490 | Yes | ||
| 45 | MARK3 | 2732 | 1.417 | 0.3515 | Yes | ||
| 46 | STRN4 | 3187 | 1.260 | 0.3329 | No | ||
| 47 | ARNT | 3640 | 1.113 | 0.3136 | No | ||
| 48 | TSC1 | 3795 | 1.062 | 0.3103 | No | ||
| 49 | FKRP | 3895 | 1.029 | 0.3097 | No | ||
| 50 | PTMA | 4082 | 0.981 | 0.3043 | No | ||
| 51 | WDR13 | 4337 | 0.917 | 0.2948 | No | ||
| 52 | ATF1 | 4778 | 0.801 | 0.2748 | No | ||
| 53 | SLC39A7 | 4783 | 0.799 | 0.2783 | No | ||
| 54 | EEF1A1 | 4797 | 0.797 | 0.2813 | No | ||
| 55 | PIAS3 | 4897 | 0.772 | 0.2796 | No | ||
| 56 | TIAL1 | 4958 | 0.756 | 0.2799 | No | ||
| 57 | ZFP37 | 4991 | 0.751 | 0.2817 | No | ||
| 58 | SMCR8 | 5065 | 0.732 | 0.2812 | No | ||
| 59 | EIF5 | 5147 | 0.712 | 0.2802 | No | ||
| 60 | CHFR | 5439 | 0.643 | 0.2674 | No | ||
| 61 | PNRC2 | 5578 | 0.615 | 0.2628 | No | ||
| 62 | NFYC | 5678 | 0.596 | 0.2603 | No | ||
| 63 | SNAP25 | 5924 | 0.543 | 0.2496 | No | ||
| 64 | UPF3B | 5953 | 0.534 | 0.2506 | No | ||
| 65 | ATP5A1 | 6077 | 0.508 | 0.2463 | No | ||
| 66 | HIF1A | 7018 | 0.299 | 0.1968 | No | ||
| 67 | RIMS1 | 7023 | 0.299 | 0.1980 | No | ||
| 68 | CCNK | 7532 | 0.195 | 0.1714 | No | ||
| 69 | LYPLA2 | 7792 | 0.149 | 0.1581 | No | ||
| 70 | IER2 | 8125 | 0.086 | 0.1405 | No | ||
| 71 | RPN1 | 8255 | 0.065 | 0.1338 | No | ||
| 72 | MAP3K4 | 8409 | 0.032 | 0.1257 | No | ||
| 73 | RNF26 | 8485 | 0.012 | 0.1217 | No | ||
| 74 | ENSA | 8509 | 0.007 | 0.1205 | No | ||
| 75 | RECK | 8677 | -0.027 | 0.1116 | No | ||
| 76 | VAX1 | 8736 | -0.038 | 0.1086 | No | ||
| 77 | BTF3 | 8800 | -0.049 | 0.1054 | No | ||
| 78 | MNT | 8801 | -0.050 | 0.1057 | No | ||
| 79 | TIA1 | 9291 | -0.157 | 0.0799 | No | ||
| 80 | POLR2A | 9330 | -0.165 | 0.0787 | No | ||
| 81 | PPP1R15B | 9411 | -0.180 | 0.0752 | No | ||
| 82 | ALG12 | 9441 | -0.186 | 0.0745 | No | ||
| 83 | TOP3A | 9904 | -0.286 | 0.0508 | No | ||
| 84 | ZBTB4 | 9927 | -0.291 | 0.0510 | No | ||
| 85 | ARF1 | 9946 | -0.295 | 0.0514 | No | ||
| 86 | PMF1 | 10000 | -0.308 | 0.0500 | No | ||
| 87 | SLC26A6 | 10009 | -0.311 | 0.0510 | No | ||
| 88 | BTRC | 10083 | -0.328 | 0.0486 | No | ||
| 89 | MPDU1 | 10231 | -0.359 | 0.0423 | No | ||
| 90 | BOP1 | 10421 | -0.399 | 0.0339 | No | ||
| 91 | ELAVL4 | 10424 | -0.400 | 0.0357 | No | ||
| 92 | RPL35A | 10779 | -0.471 | 0.0188 | No | ||
| 93 | RAD21 | 10861 | -0.486 | 0.0167 | No | ||
| 94 | ECH1 | 11139 | -0.542 | 0.0042 | No | ||
| 95 | PIGL | 11287 | -0.572 | -0.0011 | No | ||
| 96 | BAMBI | 11304 | -0.575 | 0.0008 | No | ||
| 97 | SFRS10 | 11569 | -0.627 | -0.0106 | No | ||
| 98 | NDRG3 | 11834 | -0.678 | -0.0217 | No | ||
| 99 | EIF4A1 | 12050 | -0.714 | -0.0300 | No | ||
| 100 | TAF6 | 12063 | -0.717 | -0.0273 | No | ||
| 101 | OSBPL11 | 12246 | -0.759 | -0.0335 | No | ||
| 102 | ATP5G2 | 12328 | -0.777 | -0.0343 | No | ||
| 103 | SMYD5 | 12747 | -0.870 | -0.0528 | No | ||
| 104 | ETV1 | 12869 | -0.893 | -0.0552 | No | ||
| 105 | CSNK1A1 | 12887 | -0.897 | -0.0519 | No | ||
| 106 | NACA | 13392 | -1.006 | -0.0744 | No | ||
| 107 | SLC25A19 | 13607 | -1.057 | -0.0811 | No | ||
| 108 | CELSR3 | 13978 | -1.143 | -0.0957 | No | ||
| 109 | ARCN1 | 14078 | -1.164 | -0.0956 | No | ||
| 110 | SYT11 | 14127 | -1.177 | -0.0927 | No | ||
| 111 | RPS8 | 14268 | -1.211 | -0.0946 | No | ||
| 112 | PEG3 | 14328 | -1.227 | -0.0920 | No | ||
| 113 | CACNB2 | 14846 | -1.348 | -0.1137 | No | ||
| 114 | ATP5F1 | 14860 | -1.351 | -0.1080 | No | ||
| 115 | UQCRH | 14927 | -1.368 | -0.1052 | No | ||
| 116 | UTX | 15186 | -1.435 | -0.1124 | No | ||
| 117 | MAEA | 15209 | -1.440 | -0.1068 | No | ||
| 118 | APBB1 | 15298 | -1.466 | -0.1047 | No | ||
| 119 | HOXC6 | 15488 | -1.529 | -0.1078 | No | ||
| 120 | RAB5A | 15930 | -1.673 | -0.1238 | No | ||
| 121 | B3GALT6 | 15984 | -1.692 | -0.1187 | No | ||
| 122 | SNURF | 15996 | -1.694 | -0.1113 | No | ||
| 123 | PBX3 | 16043 | -1.713 | -0.1058 | No | ||
| 124 | MORF4L2 | 16074 | -1.725 | -0.0993 | No | ||
| 125 | MAPT | 16260 | -1.784 | -0.1009 | No | ||
| 126 | ZDHHC5 | 16275 | -1.790 | -0.0933 | No | ||
| 127 | WBSCR16 | 16427 | -1.842 | -0.0928 | No | ||
| 128 | TAX1BP3 | 16586 | -1.911 | -0.0924 | No | ||
| 129 | FZD8 | 16657 | -1.944 | -0.0871 | No | ||
| 130 | ATP5H | 16663 | -1.947 | -0.0782 | No | ||
| 131 | HOXC4 | 16696 | -1.962 | -0.0707 | No | ||
| 132 | MYNN | 16776 | -1.991 | -0.0656 | No | ||
| 133 | TARDBP | 16868 | -2.043 | -0.0610 | No | ||
| 134 | WDR34 | 16911 | -2.062 | -0.0536 | No | ||
| 135 | UBE2J2 | 16985 | -2.101 | -0.0476 | No | ||
| 136 | NCDN | 17167 | -2.207 | -0.0471 | No | ||
| 137 | ZFX | 17376 | -2.347 | -0.0473 | No | ||
| 138 | RALA | 17413 | -2.375 | -0.0381 | No | ||
| 139 | DEPDC6 | 17629 | -2.532 | -0.0379 | No | ||
| 140 | PSMB5 | 17668 | -2.567 | -0.0279 | No | ||
| 141 | CIRH1A | 17672 | -2.572 | -0.0159 | No | ||
| 142 | THAP1 | 17760 | -2.646 | -0.0082 | No | ||
| 143 | DAP3 | 17812 | -2.687 | 0.0016 | No | ||
| 144 | PCBP4 | 18443 | -3.800 | -0.0146 | No | ||
| 145 | ADK | 18578 | -5.094 | 0.0021 | No |
