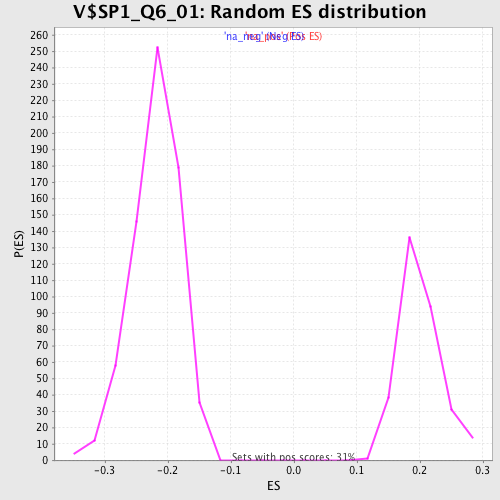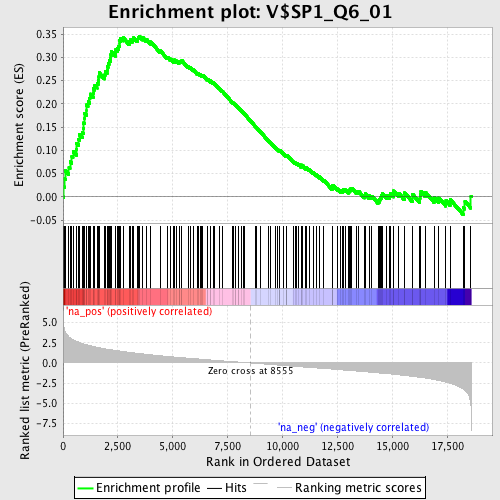
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$SP1_Q6_01 |
| Enrichment Score (ES) | 0.3457048 |
| Normalized Enrichment Score (NES) | 1.73461 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0707447 |
| FWER p-Value | 0.329 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MYOHD1 | 21 | 4.759 | 0.0223 | Yes | ||
| 2 | ABCC1 | 81 | 3.954 | 0.0386 | Yes | ||
| 3 | MAP3K11 | 104 | 3.838 | 0.0564 | Yes | ||
| 4 | PITPNM1 | 267 | 3.215 | 0.0635 | Yes | ||
| 5 | PIP5K2B | 315 | 3.124 | 0.0763 | Yes | ||
| 6 | PAK4 | 401 | 2.958 | 0.0863 | Yes | ||
| 7 | NFAT5 | 451 | 2.890 | 0.0979 | Yes | ||
| 8 | DNAJC4 | 606 | 2.693 | 0.1029 | Yes | ||
| 9 | IQGAP1 | 617 | 2.684 | 0.1156 | Yes | ||
| 10 | POLR3E | 703 | 2.581 | 0.1237 | Yes | ||
| 11 | SIPA1 | 738 | 2.534 | 0.1343 | Yes | ||
| 12 | CAMK2G | 879 | 2.414 | 0.1387 | Yes | ||
| 13 | GRB2 | 924 | 2.371 | 0.1480 | Yes | ||
| 14 | GCN5L2 | 936 | 2.361 | 0.1590 | Yes | ||
| 15 | KLF2 | 966 | 2.341 | 0.1690 | Yes | ||
| 16 | PVRL2 | 974 | 2.337 | 0.1802 | Yes | ||
| 17 | RABEP2 | 1061 | 2.280 | 0.1867 | Yes | ||
| 18 | SCYL1 | 1065 | 2.273 | 0.1978 | Yes | ||
| 19 | MAP4K2 | 1136 | 2.202 | 0.2049 | Yes | ||
| 20 | PIGW | 1208 | 2.152 | 0.2116 | Yes | ||
| 21 | TMEM24 | 1226 | 2.135 | 0.2212 | Yes | ||
| 22 | ATP2A2 | 1370 | 2.038 | 0.2235 | Yes | ||
| 23 | RHOG | 1380 | 2.031 | 0.2331 | Yes | ||
| 24 | TCF4 | 1445 | 1.998 | 0.2395 | Yes | ||
| 25 | BCL9L | 1547 | 1.941 | 0.2436 | Yes | ||
| 26 | SENP1 | 1596 | 1.913 | 0.2504 | Yes | ||
| 27 | WNT2B | 1630 | 1.893 | 0.2580 | Yes | ||
| 28 | ANP32A | 1636 | 1.892 | 0.2670 | Yes | ||
| 29 | PTPN2 | 1887 | 1.766 | 0.2622 | Yes | ||
| 30 | UPF2 | 1913 | 1.752 | 0.2695 | Yes | ||
| 31 | ELL | 2005 | 1.710 | 0.2730 | Yes | ||
| 32 | GOLGA3 | 2014 | 1.707 | 0.2810 | Yes | ||
| 33 | TLX2 | 2050 | 1.693 | 0.2874 | Yes | ||
| 34 | LDB1 | 2095 | 1.676 | 0.2933 | Yes | ||
| 35 | LASP1 | 2155 | 1.648 | 0.2982 | Yes | ||
| 36 | MYO1C | 2174 | 1.640 | 0.3053 | Yes | ||
| 37 | A2BP1 | 2193 | 1.632 | 0.3124 | Yes | ||
| 38 | ULK1 | 2377 | 1.550 | 0.3101 | Yes | ||
| 39 | AP1GBP1 | 2404 | 1.539 | 0.3163 | Yes | ||
| 40 | VAMP2 | 2485 | 1.509 | 0.3194 | Yes | ||
| 41 | KTN1 | 2534 | 1.493 | 0.3242 | Yes | ||
| 42 | KCNQ4 | 2563 | 1.483 | 0.3300 | Yes | ||
| 43 | BCL2L2 | 2566 | 1.481 | 0.3372 | Yes | ||
| 44 | SMARCD1 | 2619 | 1.457 | 0.3416 | Yes | ||
| 45 | ANKRD25 | 2738 | 1.415 | 0.3421 | Yes | ||
| 46 | TIMELESS | 3013 | 1.315 | 0.3338 | Yes | ||
| 47 | ADAM17 | 3057 | 1.304 | 0.3379 | Yes | ||
| 48 | MAZ | 3151 | 1.273 | 0.3391 | Yes | ||
| 49 | PLEKHM1 | 3211 | 1.250 | 0.3421 | Yes | ||
| 50 | ECE1 | 3377 | 1.198 | 0.3391 | Yes | ||
| 51 | CHKA | 3413 | 1.188 | 0.3430 | Yes | ||
| 52 | SNAPAP | 3471 | 1.168 | 0.3457 | Yes | ||
| 53 | RELA | 3628 | 1.116 | 0.3428 | No | ||
| 54 | EDG8 | 3786 | 1.066 | 0.3395 | No | ||
| 55 | SUV39H2 | 3992 | 1.006 | 0.3334 | No | ||
| 56 | SEC24C | 4428 | 0.890 | 0.3142 | No | ||
| 57 | DUOX1 | 4755 | 0.806 | 0.3005 | No | ||
| 58 | PIAS3 | 4897 | 0.772 | 0.2967 | No | ||
| 59 | DGKA | 5048 | 0.736 | 0.2922 | No | ||
| 60 | EPS8L2 | 5071 | 0.730 | 0.2946 | No | ||
| 61 | NTF5 | 5176 | 0.708 | 0.2924 | No | ||
| 62 | CNNM4 | 5281 | 0.681 | 0.2901 | No | ||
| 63 | PER1 | 5326 | 0.668 | 0.2911 | No | ||
| 64 | HOXC13 | 5383 | 0.655 | 0.2913 | No | ||
| 65 | CXCL12 | 5412 | 0.649 | 0.2929 | No | ||
| 66 | TEAD2 | 5695 | 0.593 | 0.2806 | No | ||
| 67 | SPATA6 | 5793 | 0.572 | 0.2782 | No | ||
| 68 | SMARCE1 | 5932 | 0.541 | 0.2733 | No | ||
| 69 | SLC24A6 | 6123 | 0.495 | 0.2655 | No | ||
| 70 | GLTP | 6165 | 0.488 | 0.2657 | No | ||
| 71 | CYP26B1 | 6264 | 0.468 | 0.2627 | No | ||
| 72 | CACNA1A | 6326 | 0.454 | 0.2616 | No | ||
| 73 | MAP3K6 | 6361 | 0.443 | 0.2620 | No | ||
| 74 | XPR1 | 6560 | 0.399 | 0.2532 | No | ||
| 75 | CHAT | 6596 | 0.393 | 0.2532 | No | ||
| 76 | NLK | 6725 | 0.363 | 0.2481 | No | ||
| 77 | LOXL4 | 6730 | 0.362 | 0.2497 | No | ||
| 78 | CSNK1D | 6835 | 0.340 | 0.2457 | No | ||
| 79 | FOSB | 6919 | 0.321 | 0.2428 | No | ||
| 80 | KLF5 | 7111 | 0.280 | 0.2338 | No | ||
| 81 | CDK5R1 | 7253 | 0.251 | 0.2274 | No | ||
| 82 | RAB35 | 7728 | 0.161 | 0.2025 | No | ||
| 83 | PGM2L1 | 7752 | 0.156 | 0.2021 | No | ||
| 84 | APLP1 | 7839 | 0.139 | 0.1981 | No | ||
| 85 | PEX14 | 7853 | 0.136 | 0.1980 | No | ||
| 86 | SLC30A3 | 7975 | 0.113 | 0.1920 | No | ||
| 87 | FGF11 | 8002 | 0.108 | 0.1912 | No | ||
| 88 | SLC35F5 | 8120 | 0.088 | 0.1853 | No | ||
| 89 | PGF | 8216 | 0.070 | 0.1805 | No | ||
| 90 | CALM3 | 8250 | 0.066 | 0.1790 | No | ||
| 91 | ADCK1 | 8775 | -0.045 | 0.1508 | No | ||
| 92 | TGFB1 | 8803 | -0.050 | 0.1496 | No | ||
| 93 | BRI3BP | 9005 | -0.092 | 0.1392 | No | ||
| 94 | CPD | 9366 | -0.171 | 0.1205 | No | ||
| 95 | NFYB | 9430 | -0.183 | 0.1180 | No | ||
| 96 | ACBD5 | 9673 | -0.236 | 0.1060 | No | ||
| 97 | NOTCH3 | 9775 | -0.261 | 0.1019 | No | ||
| 98 | NXPH4 | 9848 | -0.275 | 0.0993 | No | ||
| 99 | JUB | 9857 | -0.276 | 0.1002 | No | ||
| 100 | TAGLN2 | 9883 | -0.280 | 0.1003 | No | ||
| 101 | EFNB3 | 10036 | -0.316 | 0.0936 | No | ||
| 102 | RAB26 | 10162 | -0.342 | 0.0885 | No | ||
| 103 | HES7 | 10166 | -0.342 | 0.0900 | No | ||
| 104 | FKBP8 | 10200 | -0.351 | 0.0900 | No | ||
| 105 | EFNA3 | 10508 | -0.413 | 0.0754 | No | ||
| 106 | STMN1 | 10588 | -0.431 | 0.0732 | No | ||
| 107 | SLC2A1 | 10655 | -0.443 | 0.0718 | No | ||
| 108 | ASCL2 | 10746 | -0.464 | 0.0692 | No | ||
| 109 | PIAS1 | 10846 | -0.483 | 0.0663 | No | ||
| 110 | HTATIP | 10847 | -0.484 | 0.0687 | No | ||
| 111 | APLP2 | 10912 | -0.497 | 0.0676 | No | ||
| 112 | PIGN | 11062 | -0.530 | 0.0622 | No | ||
| 113 | INPPL1 | 11096 | -0.536 | 0.0630 | No | ||
| 114 | CORO1C | 11232 | -0.561 | 0.0585 | No | ||
| 115 | SLC1A2 | 11394 | -0.593 | 0.0527 | No | ||
| 116 | CSPG4 | 11540 | -0.620 | 0.0479 | No | ||
| 117 | OTX1 | 11683 | -0.647 | 0.0434 | No | ||
| 118 | MAP2K7 | 11878 | -0.685 | 0.0363 | No | ||
| 119 | PHCA | 12262 | -0.761 | 0.0193 | No | ||
| 120 | DRAP1 | 12271 | -0.762 | 0.0226 | No | ||
| 121 | GABARAPL2 | 12290 | -0.768 | 0.0254 | No | ||
| 122 | GAS7 | 12516 | -0.816 | 0.0172 | No | ||
| 123 | TAGLN | 12664 | -0.850 | 0.0135 | No | ||
| 124 | CDC37 | 12739 | -0.867 | 0.0137 | No | ||
| 125 | CRY1 | 12790 | -0.877 | 0.0153 | No | ||
| 126 | LZTS2 | 12876 | -0.894 | 0.0152 | No | ||
| 127 | BCL7C | 13008 | -0.924 | 0.0126 | No | ||
| 128 | TRIM28 | 13051 | -0.932 | 0.0149 | No | ||
| 129 | PTOV1 | 13096 | -0.941 | 0.0172 | No | ||
| 130 | GGN | 13161 | -0.955 | 0.0184 | No | ||
| 131 | ELAC2 | 13367 | -1.002 | 0.0123 | No | ||
| 132 | HTR7 | 13474 | -1.025 | 0.0116 | No | ||
| 133 | PHOX2A | 13734 | -1.085 | 0.0029 | No | ||
| 134 | RELB | 13763 | -1.089 | 0.0068 | No | ||
| 135 | CLPTM1 | 13947 | -1.136 | 0.0024 | No | ||
| 136 | UGP2 | 14074 | -1.163 | 0.0014 | No | ||
| 137 | HCN4 | 14354 | -1.233 | -0.0077 | No | ||
| 138 | PTCH2 | 14422 | -1.252 | -0.0051 | No | ||
| 139 | EPS15 | 14453 | -1.258 | -0.0006 | No | ||
| 140 | NET1 | 14502 | -1.270 | 0.0031 | No | ||
| 141 | CBLN1 | 14547 | -1.281 | 0.0070 | No | ||
| 142 | IRX3 | 14731 | -1.321 | 0.0036 | No | ||
| 143 | MAPK7 | 14872 | -1.355 | 0.0027 | No | ||
| 144 | RAB2B | 14904 | -1.362 | 0.0078 | No | ||
| 145 | ELAVL3 | 15054 | -1.401 | 0.0066 | No | ||
| 146 | SFRS2 | 15068 | -1.403 | 0.0128 | No | ||
| 147 | PARD6A | 15307 | -1.469 | 0.0072 | No | ||
| 148 | FDX1 | 15562 | -1.551 | 0.0011 | No | ||
| 149 | NAV1 | 15565 | -1.551 | 0.0086 | No | ||
| 150 | PSMC6 | 15920 | -1.669 | -0.0023 | No | ||
| 151 | DDB1 | 15921 | -1.670 | 0.0059 | No | ||
| 152 | IDH3A | 16264 | -1.785 | -0.0038 | No | ||
| 153 | ROM1 | 16269 | -1.787 | 0.0048 | No | ||
| 154 | SLC18A3 | 16307 | -1.802 | 0.0117 | No | ||
| 155 | OAZ2 | 16520 | -1.882 | 0.0094 | No | ||
| 156 | POLR2I | 16908 | -2.061 | -0.0014 | No | ||
| 157 | GPR3 | 17125 | -2.179 | -0.0023 | No | ||
| 158 | ADAM15 | 17449 | -2.398 | -0.0080 | No | ||
| 159 | VASP | 17638 | -2.545 | -0.0056 | No | ||
| 160 | SHMT1 | 18234 | -3.231 | -0.0219 | No | ||
| 161 | ACE | 18315 | -3.409 | -0.0094 | No | ||
| 162 | SMOC1 | 18588 | -5.208 | 0.0015 | No |