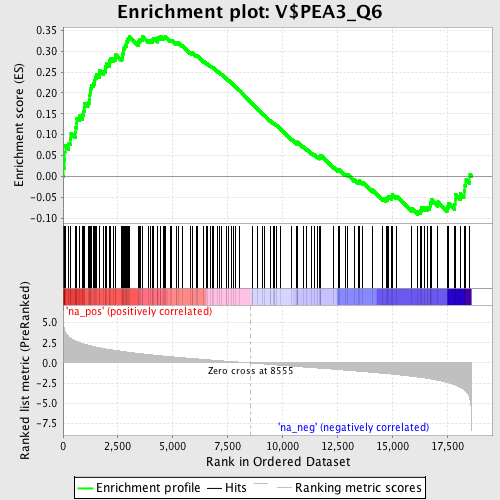
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$PEA3_Q6 |
| Enrichment Score (ES) | 0.33595324 |
| Normalized Enrichment Score (NES) | 1.6799587 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.06282526 |
| FWER p-Value | 0.462 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TRIM8 | 19 | 4.785 | 0.0212 | Yes | ||
| 2 | KCNAB2 | 45 | 4.335 | 0.0399 | Yes | ||
| 3 | HHEX | 72 | 3.995 | 0.0571 | Yes | ||
| 4 | SLC35C1 | 87 | 3.910 | 0.0745 | Yes | ||
| 5 | RPS6KA4 | 265 | 3.219 | 0.0798 | Yes | ||
| 6 | PTPN6 | 332 | 3.080 | 0.0905 | Yes | ||
| 7 | MARK2 | 357 | 3.024 | 0.1033 | Yes | ||
| 8 | LRMP | 566 | 2.734 | 0.1047 | Yes | ||
| 9 | DCLRE1C | 574 | 2.725 | 0.1169 | Yes | ||
| 10 | EPC1 | 609 | 2.691 | 0.1276 | Yes | ||
| 11 | ADRBK1 | 627 | 2.663 | 0.1390 | Yes | ||
| 12 | SIPA1 | 738 | 2.534 | 0.1448 | Yes | ||
| 13 | IL18BP | 885 | 2.408 | 0.1481 | Yes | ||
| 14 | SRRM1 | 930 | 2.365 | 0.1567 | Yes | ||
| 15 | DGKZ | 965 | 2.341 | 0.1657 | Yes | ||
| 16 | CENTD2 | 981 | 2.334 | 0.1757 | Yes | ||
| 17 | MAP4K2 | 1136 | 2.202 | 0.1776 | Yes | ||
| 18 | NUMA1 | 1186 | 2.165 | 0.1850 | Yes | ||
| 19 | ANKRD1 | 1215 | 2.143 | 0.1934 | Yes | ||
| 20 | TMEM24 | 1226 | 2.135 | 0.2028 | Yes | ||
| 21 | TNNI2 | 1263 | 2.112 | 0.2106 | Yes | ||
| 22 | WASF2 | 1291 | 2.090 | 0.2189 | Yes | ||
| 23 | RHOG | 1380 | 2.031 | 0.2235 | Yes | ||
| 24 | PICALM | 1442 | 1.999 | 0.2295 | Yes | ||
| 25 | POU2AF1 | 1454 | 1.996 | 0.2382 | Yes | ||
| 26 | GTF2H1 | 1526 | 1.955 | 0.2434 | Yes | ||
| 27 | TCIRG1 | 1644 | 1.887 | 0.2458 | Yes | ||
| 28 | PLCB3 | 1662 | 1.879 | 0.2536 | Yes | ||
| 29 | TNFRSF19L | 1844 | 1.787 | 0.2521 | Yes | ||
| 30 | ADAMTS4 | 1920 | 1.747 | 0.2561 | Yes | ||
| 31 | CDC14A | 1936 | 1.740 | 0.2634 | Yes | ||
| 32 | BLNK | 1972 | 1.721 | 0.2695 | Yes | ||
| 33 | PUM1 | 2103 | 1.671 | 0.2702 | Yes | ||
| 34 | ESRRA | 2115 | 1.665 | 0.2773 | Yes | ||
| 35 | CAMK1D | 2178 | 1.638 | 0.2816 | Yes | ||
| 36 | CPNE8 | 2302 | 1.584 | 0.2823 | Yes | ||
| 37 | RNASEL | 2372 | 1.554 | 0.2857 | Yes | ||
| 38 | XPNPEP1 | 2401 | 1.539 | 0.2914 | Yes | ||
| 39 | NCAM1 | 2678 | 1.435 | 0.2831 | Yes | ||
| 40 | SORL1 | 2718 | 1.423 | 0.2876 | Yes | ||
| 41 | EHF | 2723 | 1.420 | 0.2939 | Yes | ||
| 42 | PBXIP1 | 2741 | 1.414 | 0.2996 | Yes | ||
| 43 | CAP1 | 2752 | 1.409 | 0.3056 | Yes | ||
| 44 | PRKACB | 2804 | 1.387 | 0.3092 | Yes | ||
| 45 | LAG3 | 2822 | 1.380 | 0.3147 | Yes | ||
| 46 | RUNX3 | 2903 | 1.349 | 0.3166 | Yes | ||
| 47 | NCF2 | 2907 | 1.348 | 0.3227 | Yes | ||
| 48 | GPD1 | 2935 | 1.339 | 0.3275 | Yes | ||
| 49 | IPO7 | 2987 | 1.323 | 0.3309 | Yes | ||
| 50 | UNC93B1 | 3008 | 1.317 | 0.3359 | Yes | ||
| 51 | FUCA1 | 3431 | 1.181 | 0.3185 | Yes | ||
| 52 | PTPRC | 3435 | 1.179 | 0.3238 | Yes | ||
| 53 | POLD4 | 3481 | 1.165 | 0.3268 | Yes | ||
| 54 | ITPKB | 3542 | 1.145 | 0.3289 | Yes | ||
| 55 | CUGBP1 | 3616 | 1.119 | 0.3301 | Yes | ||
| 56 | TRIM33 | 3626 | 1.116 | 0.3348 | Yes | ||
| 57 | RIN1 | 3886 | 1.032 | 0.3255 | Yes | ||
| 58 | SNCG | 3991 | 1.006 | 0.3246 | Yes | ||
| 59 | ARHGDIB | 4075 | 0.983 | 0.3246 | Yes | ||
| 60 | CBL | 4089 | 0.980 | 0.3285 | Yes | ||
| 61 | ELK4 | 4121 | 0.969 | 0.3313 | Yes | ||
| 62 | SV2A | 4298 | 0.927 | 0.3261 | Yes | ||
| 63 | MFN2 | 4302 | 0.925 | 0.3302 | Yes | ||
| 64 | FLI1 | 4323 | 0.920 | 0.3334 | Yes | ||
| 65 | SEC24C | 4428 | 0.890 | 0.3319 | Yes | ||
| 66 | CD84 | 4446 | 0.884 | 0.3351 | Yes | ||
| 67 | ZRANB1 | 4578 | 0.851 | 0.3319 | Yes | ||
| 68 | ETV3 | 4631 | 0.839 | 0.3330 | Yes | ||
| 69 | SELL | 4649 | 0.837 | 0.3360 | Yes | ||
| 70 | CD5 | 4891 | 0.773 | 0.3265 | No | ||
| 71 | TIAL1 | 4958 | 0.756 | 0.3264 | No | ||
| 72 | CAPZA1 | 5149 | 0.712 | 0.3194 | No | ||
| 73 | FUT11 | 5186 | 0.705 | 0.3208 | No | ||
| 74 | LAMC1 | 5264 | 0.685 | 0.3198 | No | ||
| 75 | PIK4CB | 5423 | 0.646 | 0.3142 | No | ||
| 76 | SPATA6 | 5793 | 0.572 | 0.2969 | No | ||
| 77 | IRAK4 | 5892 | 0.551 | 0.2941 | No | ||
| 78 | RAB39 | 5905 | 0.547 | 0.2960 | No | ||
| 79 | IVL | 6058 | 0.512 | 0.2901 | No | ||
| 80 | FCGR3A | 6130 | 0.494 | 0.2886 | No | ||
| 81 | CTSW | 6420 | 0.430 | 0.2749 | No | ||
| 82 | FEN1 | 6554 | 0.401 | 0.2696 | No | ||
| 83 | CD53 | 6570 | 0.398 | 0.2706 | No | ||
| 84 | TCEB3 | 6727 | 0.363 | 0.2638 | No | ||
| 85 | VCAM1 | 6798 | 0.348 | 0.2617 | No | ||
| 86 | CTSS | 6861 | 0.334 | 0.2599 | No | ||
| 87 | CACNA1E | 7047 | 0.294 | 0.2512 | No | ||
| 88 | LPXN | 7124 | 0.277 | 0.2484 | No | ||
| 89 | PRF1 | 7206 | 0.260 | 0.2452 | No | ||
| 90 | ARF3 | 7429 | 0.213 | 0.2341 | No | ||
| 91 | SYTL1 | 7459 | 0.206 | 0.2335 | No | ||
| 92 | LRFN4 | 7558 | 0.191 | 0.2291 | No | ||
| 93 | ITPR2 | 7687 | 0.169 | 0.2230 | No | ||
| 94 | PGM2L1 | 7752 | 0.156 | 0.2202 | No | ||
| 95 | RPS3 | 7868 | 0.133 | 0.2146 | No | ||
| 96 | XCL1 | 8030 | 0.104 | 0.2064 | No | ||
| 97 | IL18 | 8619 | -0.015 | 0.1746 | No | ||
| 98 | GRIN2B | 8638 | -0.020 | 0.1737 | No | ||
| 99 | GPA33 | 8856 | -0.061 | 0.1622 | No | ||
| 100 | MR1 | 9077 | -0.108 | 0.1508 | No | ||
| 101 | LIN28 | 9180 | -0.130 | 0.1459 | No | ||
| 102 | CTBP2 | 9431 | -0.183 | 0.1332 | No | ||
| 103 | GFI1 | 9461 | -0.191 | 0.1325 | No | ||
| 104 | MMRN2 | 9591 | -0.218 | 0.1265 | No | ||
| 105 | SPI1 | 9616 | -0.224 | 0.1263 | No | ||
| 106 | GAB2 | 9649 | -0.231 | 0.1256 | No | ||
| 107 | HIPK1 | 9732 | -0.249 | 0.1223 | No | ||
| 108 | DMBT1 | 9885 | -0.281 | 0.1154 | No | ||
| 109 | ELAVL4 | 10424 | -0.400 | 0.0881 | No | ||
| 110 | SLC41A1 | 10630 | -0.438 | 0.0790 | No | ||
| 111 | CDKN2C | 10668 | -0.447 | 0.0791 | No | ||
| 112 | MAPKAPK2 | 10672 | -0.448 | 0.0810 | No | ||
| 113 | KAZALD1 | 10683 | -0.449 | 0.0825 | No | ||
| 114 | CD3E | 10934 | -0.503 | 0.0713 | No | ||
| 115 | INPPL1 | 11096 | -0.536 | 0.0651 | No | ||
| 116 | CYR61 | 11314 | -0.578 | 0.0560 | No | ||
| 117 | PAX6 | 11434 | -0.603 | 0.0524 | No | ||
| 118 | ELOVL1 | 11589 | -0.629 | 0.0469 | No | ||
| 119 | FGF23 | 11688 | -0.648 | 0.0446 | No | ||
| 120 | MMP7 | 11715 | -0.654 | 0.0463 | No | ||
| 121 | CD69 | 11724 | -0.656 | 0.0489 | No | ||
| 122 | VPS11 | 11742 | -0.658 | 0.0510 | No | ||
| 123 | TYSND1 | 12341 | -0.780 | 0.0222 | No | ||
| 124 | EPHA2 | 12535 | -0.820 | 0.0156 | No | ||
| 125 | SLC2A3 | 12577 | -0.831 | 0.0172 | No | ||
| 126 | LZTS2 | 12876 | -0.894 | 0.0052 | No | ||
| 127 | SPRR4 | 12958 | -0.911 | 0.0051 | No | ||
| 128 | DPP3 | 13276 | -0.981 | -0.0076 | No | ||
| 129 | SELP | 13465 | -1.022 | -0.0130 | No | ||
| 130 | SSBP3 | 13506 | -1.031 | -0.0104 | No | ||
| 131 | NRGN | 13666 | -1.067 | -0.0141 | No | ||
| 132 | IL2RA | 14100 | -1.170 | -0.0321 | No | ||
| 133 | BDNF | 14578 | -1.287 | -0.0520 | No | ||
| 134 | BCL2L14 | 14717 | -1.317 | -0.0533 | No | ||
| 135 | GATA3 | 14786 | -1.333 | -0.0508 | No | ||
| 136 | PDGFD | 14833 | -1.345 | -0.0471 | No | ||
| 137 | LTBR | 14978 | -1.385 | -0.0484 | No | ||
| 138 | ST7L | 15015 | -1.393 | -0.0439 | No | ||
| 139 | VDAC2 | 15201 | -1.438 | -0.0473 | No | ||
| 140 | ABLIM1 | 15889 | -1.658 | -0.0768 | No | ||
| 141 | LALBA | 16156 | -1.753 | -0.0831 | No | ||
| 142 | ZDHHC5 | 16275 | -1.790 | -0.0812 | No | ||
| 143 | CMAS | 16320 | -1.806 | -0.0752 | No | ||
| 144 | MARK1 | 16453 | -1.851 | -0.0737 | No | ||
| 145 | ANK3 | 16615 | -1.925 | -0.0735 | No | ||
| 146 | GPR120 | 16721 | -1.972 | -0.0700 | No | ||
| 147 | PRDX6 | 16743 | -1.981 | -0.0620 | No | ||
| 148 | ZNF289 | 16792 | -2.000 | -0.0553 | No | ||
| 149 | NDUFS2 | 17085 | -2.153 | -0.0611 | No | ||
| 150 | EXTL2 | 17499 | -2.421 | -0.0723 | No | ||
| 151 | PAFAH2 | 17566 | -2.470 | -0.0644 | No | ||
| 152 | LCK | 17843 | -2.721 | -0.0667 | No | ||
| 153 | SLC26A9 | 17864 | -2.740 | -0.0551 | No | ||
| 154 | KCNQ1 | 17892 | -2.761 | -0.0437 | No | ||
| 155 | SLC30A7 | 18106 | -3.038 | -0.0412 | No | ||
| 156 | AKT3 | 18272 | -3.301 | -0.0348 | No | ||
| 157 | NT5C2 | 18314 | -3.409 | -0.0212 | No | ||
| 158 | TAF5 | 18361 | -3.557 | -0.0072 | No | ||
| 159 | SLAMF9 | 18543 | -4.514 | 0.0040 | No |
