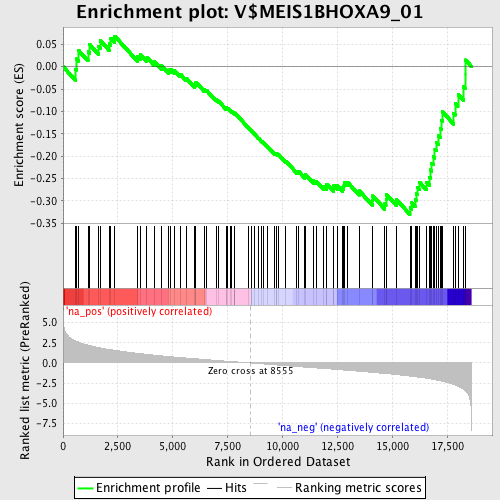
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$MEIS1BHOXA9_01 |
| Enrichment Score (ES) | -0.33058083 |
| Normalized Enrichment Score (NES) | -1.4094098 |
| Nominal p-value | 0.02511774 |
| FDR q-value | 0.5300049 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PDLIM1 | 568 | 2.733 | -0.0053 | No | ||
| 2 | ATBF1 | 611 | 2.691 | 0.0174 | No | ||
| 3 | NFKB2 | 704 | 2.580 | 0.0363 | No | ||
| 4 | PIK3CG | 1139 | 2.200 | 0.0333 | No | ||
| 5 | TBL1X | 1203 | 2.156 | 0.0499 | No | ||
| 6 | DMD | 1614 | 1.903 | 0.0454 | No | ||
| 7 | TLE4 | 1692 | 1.857 | 0.0585 | No | ||
| 8 | CSMD3 | 2108 | 1.668 | 0.0516 | No | ||
| 9 | MYO1C | 2174 | 1.640 | 0.0633 | No | ||
| 10 | MBNL1 | 2349 | 1.564 | 0.0684 | No | ||
| 11 | EIF4ENIF1 | 3401 | 1.191 | 0.0227 | No | ||
| 12 | AMOT | 3511 | 1.156 | 0.0275 | No | ||
| 13 | USH1G | 3822 | 1.051 | 0.0205 | No | ||
| 14 | PPP1R10 | 4160 | 0.960 | 0.0113 | No | ||
| 15 | PTHR1 | 4482 | 0.874 | 0.0020 | No | ||
| 16 | SYNPR | 4794 | 0.798 | -0.0074 | No | ||
| 17 | MDK | 4893 | 0.773 | -0.0055 | No | ||
| 18 | KLHL4 | 5076 | 0.728 | -0.0085 | No | ||
| 19 | OTOP2 | 5342 | 0.665 | -0.0167 | No | ||
| 20 | UNC5C | 5625 | 0.606 | -0.0263 | No | ||
| 21 | HOXB7 | 5999 | 0.523 | -0.0416 | No | ||
| 22 | UTY | 6033 | 0.517 | -0.0386 | No | ||
| 23 | KLRC2 | 6050 | 0.513 | -0.0347 | No | ||
| 24 | KLHL1 | 6421 | 0.430 | -0.0506 | No | ||
| 25 | SOST | 6521 | 0.408 | -0.0522 | No | ||
| 26 | CYLD | 6978 | 0.308 | -0.0740 | No | ||
| 27 | OTX2 | 7078 | 0.286 | -0.0767 | No | ||
| 28 | PIK3R1 | 7425 | 0.214 | -0.0934 | No | ||
| 29 | ARF3 | 7429 | 0.213 | -0.0915 | No | ||
| 30 | NOG | 7484 | 0.203 | -0.0926 | No | ||
| 31 | INSM1 | 7621 | 0.181 | -0.0982 | No | ||
| 32 | CISH | 7688 | 0.168 | -0.1002 | No | ||
| 33 | NFIL3 | 7808 | 0.146 | -0.1053 | No | ||
| 34 | HS3ST3A1 | 7821 | 0.143 | -0.1046 | No | ||
| 35 | PHOX2B | 7825 | 0.142 | -0.1035 | No | ||
| 36 | KLRC3 | 8469 | 0.016 | -0.1380 | No | ||
| 37 | EBF2 | 8590 | -0.009 | -0.1444 | No | ||
| 38 | UBR1 | 8608 | -0.013 | -0.1452 | No | ||
| 39 | CHODL | 8703 | -0.032 | -0.1500 | No | ||
| 40 | IL19 | 8893 | -0.069 | -0.1596 | No | ||
| 41 | ESRRG | 9060 | -0.103 | -0.1676 | No | ||
| 42 | TCF3 | 9110 | -0.115 | -0.1692 | No | ||
| 43 | CHN2 | 9306 | -0.160 | -0.1782 | No | ||
| 44 | NRAS | 9654 | -0.233 | -0.1948 | No | ||
| 45 | TTC12 | 9707 | -0.243 | -0.1953 | No | ||
| 46 | HIPK1 | 9732 | -0.249 | -0.1943 | No | ||
| 47 | GRM8 | 9830 | -0.271 | -0.1970 | No | ||
| 48 | ATP2B3 | 10155 | -0.340 | -0.2114 | No | ||
| 49 | PFTK1 | 10654 | -0.443 | -0.2341 | No | ||
| 50 | HOXC12 | 10737 | -0.461 | -0.2343 | No | ||
| 51 | ARPC5 | 11018 | -0.522 | -0.2446 | No | ||
| 52 | HOXD9 | 11054 | -0.529 | -0.2416 | No | ||
| 53 | CD79B | 11408 | -0.595 | -0.2551 | No | ||
| 54 | HOXA11 | 11536 | -0.620 | -0.2562 | No | ||
| 55 | SIM1 | 11874 | -0.685 | -0.2680 | No | ||
| 56 | IRS1 | 11998 | -0.706 | -0.2681 | No | ||
| 57 | ZIC5 | 12015 | -0.707 | -0.2624 | No | ||
| 58 | GABRG2 | 12334 | -0.779 | -0.2724 | No | ||
| 59 | KCNV2 | 12340 | -0.780 | -0.2654 | No | ||
| 60 | RASGRF1 | 12500 | -0.811 | -0.2665 | No | ||
| 61 | HOXA3 | 12730 | -0.867 | -0.2708 | No | ||
| 62 | DNAJB7 | 12800 | -0.879 | -0.2664 | No | ||
| 63 | ZIC4 | 12811 | -0.882 | -0.2587 | No | ||
| 64 | ACTN2 | 12960 | -0.911 | -0.2583 | No | ||
| 65 | LPHN2 | 13497 | -1.030 | -0.2777 | No | ||
| 66 | TPM2 | 14103 | -1.170 | -0.2995 | No | ||
| 67 | CLPX | 14112 | -1.172 | -0.2890 | No | ||
| 68 | DRD3 | 14666 | -1.305 | -0.3068 | No | ||
| 69 | WNT6 | 14718 | -1.318 | -0.2973 | No | ||
| 70 | IRX3 | 14731 | -1.321 | -0.2857 | No | ||
| 71 | UTX | 15186 | -1.435 | -0.2969 | No | ||
| 72 | RAB10 | 15811 | -1.629 | -0.3155 | Yes | ||
| 73 | NR2F2 | 15883 | -1.657 | -0.3039 | Yes | ||
| 74 | GBX2 | 16045 | -1.713 | -0.2967 | Yes | ||
| 75 | HOXA6 | 16102 | -1.734 | -0.2837 | Yes | ||
| 76 | CACNB3 | 16138 | -1.744 | -0.2694 | Yes | ||
| 77 | GNAO1 | 16243 | -1.778 | -0.2585 | Yes | ||
| 78 | CDH13 | 16562 | -1.899 | -0.2581 | Yes | ||
| 79 | HOXC4 | 16696 | -1.962 | -0.2470 | Yes | ||
| 80 | PCSK2 | 16737 | -1.978 | -0.2308 | Yes | ||
| 81 | FOXB1 | 16797 | -2.004 | -0.2154 | Yes | ||
| 82 | OLFM1 | 16871 | -2.045 | -0.2004 | Yes | ||
| 83 | HOXC5 | 16947 | -2.080 | -0.1852 | Yes | ||
| 84 | TBR1 | 17011 | -2.112 | -0.1690 | Yes | ||
| 85 | RPP25 | 17122 | -2.178 | -0.1547 | Yes | ||
| 86 | EDG2 | 17196 | -2.227 | -0.1380 | Yes | ||
| 87 | DSPP | 17250 | -2.259 | -0.1199 | Yes | ||
| 88 | GNB1L | 17287 | -2.280 | -0.1007 | Yes | ||
| 89 | CRX | 17805 | -2.683 | -0.1037 | Yes | ||
| 90 | TNFRSF8 | 17889 | -2.759 | -0.0826 | Yes | ||
| 91 | HNF4G | 17999 | -2.896 | -0.0616 | Yes | ||
| 92 | AP3S1 | 18254 | -3.277 | -0.0449 | Yes | ||
| 93 | COPS3 | 18336 | -3.469 | -0.0171 | Yes | ||
| 94 | MRPS18B | 18337 | -3.470 | 0.0151 | Yes |
