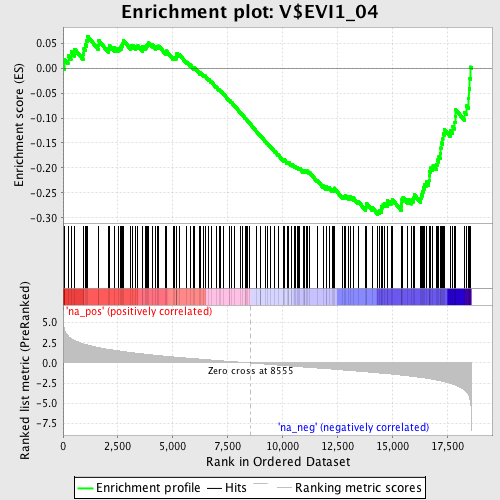
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$EVI1_04 |
| Enrichment Score (ES) | -0.29346126 |
| Normalized Enrichment Score (NES) | -1.3527704 |
| Nominal p-value | 0.020057306 |
| FDR q-value | 0.57582366 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BCL11A | 59 | 4.158 | 0.0175 | No | ||
| 2 | PPAP2B | 227 | 3.341 | 0.0252 | No | ||
| 3 | RERE | 362 | 3.014 | 0.0329 | No | ||
| 4 | GDI1 | 525 | 2.797 | 0.0381 | No | ||
| 5 | MID1 | 919 | 2.375 | 0.0287 | No | ||
| 6 | TOB1 | 948 | 2.352 | 0.0389 | No | ||
| 7 | RPS6KB1 | 1004 | 2.317 | 0.0474 | No | ||
| 8 | MAX | 1072 | 2.267 | 0.0551 | No | ||
| 9 | RAI1 | 1109 | 2.230 | 0.0643 | No | ||
| 10 | DMD | 1614 | 1.903 | 0.0465 | No | ||
| 11 | WNT2B | 1630 | 1.893 | 0.0551 | No | ||
| 12 | PTGFR | 2057 | 1.689 | 0.0404 | No | ||
| 13 | CSMD3 | 2108 | 1.668 | 0.0461 | No | ||
| 14 | MBNL1 | 2349 | 1.564 | 0.0409 | No | ||
| 15 | HNRPA0 | 2505 | 1.504 | 0.0400 | No | ||
| 16 | POU4F2 | 2593 | 1.468 | 0.0426 | No | ||
| 17 | PPOX | 2673 | 1.439 | 0.0455 | No | ||
| 18 | EHF | 2723 | 1.420 | 0.0499 | No | ||
| 19 | FOXP2 | 2739 | 1.415 | 0.0561 | No | ||
| 20 | MYH2 | 3073 | 1.299 | 0.0446 | No | ||
| 21 | HOXD4 | 3168 | 1.267 | 0.0458 | No | ||
| 22 | STOML2 | 3316 | 1.216 | 0.0439 | No | ||
| 23 | H3F3B | 3409 | 1.189 | 0.0448 | No | ||
| 24 | CPNE1 | 3620 | 1.117 | 0.0390 | No | ||
| 25 | RELA | 3628 | 1.116 | 0.0442 | No | ||
| 26 | CLASP1 | 3765 | 1.072 | 0.0422 | No | ||
| 27 | NOTCH2 | 3803 | 1.058 | 0.0455 | No | ||
| 28 | PLAG1 | 3845 | 1.044 | 0.0485 | No | ||
| 29 | ARHGAP6 | 3898 | 1.029 | 0.0508 | No | ||
| 30 | SEMA6A | 4058 | 0.987 | 0.0471 | No | ||
| 31 | HTR3B | 4227 | 0.940 | 0.0427 | No | ||
| 32 | SLC4A4 | 4290 | 0.929 | 0.0439 | No | ||
| 33 | TNFSF4 | 4363 | 0.909 | 0.0446 | No | ||
| 34 | CER1 | 4684 | 0.826 | 0.0314 | No | ||
| 35 | COX8A | 4698 | 0.822 | 0.0347 | No | ||
| 36 | CRH | 5011 | 0.745 | 0.0216 | No | ||
| 37 | PDE4D | 5085 | 0.727 | 0.0212 | No | ||
| 38 | EIF5 | 5147 | 0.712 | 0.0215 | No | ||
| 39 | EDN1 | 5173 | 0.708 | 0.0237 | No | ||
| 40 | SLC40A1 | 5181 | 0.707 | 0.0268 | No | ||
| 41 | FUT11 | 5186 | 0.705 | 0.0301 | No | ||
| 42 | AP3M2 | 5305 | 0.674 | 0.0271 | No | ||
| 43 | GSH1 | 5630 | 0.604 | 0.0125 | No | ||
| 44 | STC1 | 5791 | 0.573 | 0.0067 | No | ||
| 45 | DNAJA4 | 5960 | 0.533 | 0.0003 | No | ||
| 46 | HOXB7 | 5999 | 0.523 | 0.0008 | No | ||
| 47 | KLF7 | 6236 | 0.473 | -0.0096 | No | ||
| 48 | CABLES1 | 6265 | 0.468 | -0.0088 | No | ||
| 49 | SFRS1 | 6416 | 0.432 | -0.0148 | No | ||
| 50 | PRL | 6470 | 0.419 | -0.0156 | No | ||
| 51 | TPM1 | 6631 | 0.384 | -0.0223 | No | ||
| 52 | PDGFC | 6759 | 0.357 | -0.0274 | No | ||
| 53 | HAS2 | 6976 | 0.309 | -0.0376 | No | ||
| 54 | KLF5 | 7111 | 0.280 | -0.0434 | No | ||
| 55 | P2RY12 | 7189 | 0.263 | -0.0463 | No | ||
| 56 | PPIL5 | 7316 | 0.239 | -0.0519 | No | ||
| 57 | NT5C1B | 7566 | 0.189 | -0.0645 | No | ||
| 58 | HOXB3 | 7595 | 0.185 | -0.0651 | No | ||
| 59 | RORB | 7671 | 0.172 | -0.0683 | No | ||
| 60 | PHOX2B | 7825 | 0.142 | -0.0759 | No | ||
| 61 | OTP | 8092 | 0.092 | -0.0898 | No | ||
| 62 | LMO4 | 8183 | 0.077 | -0.0943 | No | ||
| 63 | MYH4 | 8296 | 0.058 | -0.1001 | No | ||
| 64 | ATOH1 | 8329 | 0.048 | -0.1016 | No | ||
| 65 | COL13A1 | 8370 | 0.041 | -0.1036 | No | ||
| 66 | BCL11B | 8393 | 0.035 | -0.1046 | No | ||
| 67 | TWIST1 | 8516 | 0.006 | -0.1112 | No | ||
| 68 | NRG1 | 8797 | -0.049 | -0.1261 | No | ||
| 69 | MYO3B | 8979 | -0.086 | -0.1355 | No | ||
| 70 | NEUROD2 | 8993 | -0.087 | -0.1357 | No | ||
| 71 | CRYAB | 9013 | -0.094 | -0.1363 | No | ||
| 72 | CYP46A1 | 9225 | -0.142 | -0.1470 | No | ||
| 73 | ZIC1 | 9310 | -0.161 | -0.1508 | No | ||
| 74 | ISL1 | 9457 | -0.189 | -0.1577 | No | ||
| 75 | NRAS | 9654 | -0.233 | -0.1672 | No | ||
| 76 | RCOR1 | 9798 | -0.265 | -0.1736 | No | ||
| 77 | GLRA2 | 10041 | -0.318 | -0.1852 | No | ||
| 78 | PCDHA5 | 10063 | -0.324 | -0.1847 | No | ||
| 79 | IL2RG | 10070 | -0.325 | -0.1834 | No | ||
| 80 | CASQ2 | 10222 | -0.356 | -0.1898 | No | ||
| 81 | PPARGC1A | 10259 | -0.366 | -0.1899 | No | ||
| 82 | LRRTM3 | 10283 | -0.371 | -0.1893 | No | ||
| 83 | EYA1 | 10412 | -0.397 | -0.1943 | No | ||
| 84 | ELAVL4 | 10424 | -0.400 | -0.1929 | No | ||
| 85 | LRRTM1 | 10529 | -0.418 | -0.1964 | No | ||
| 86 | NRXN3 | 10596 | -0.433 | -0.1978 | No | ||
| 87 | CDKN2C | 10668 | -0.447 | -0.1995 | No | ||
| 88 | SMPX | 10744 | -0.463 | -0.2012 | No | ||
| 89 | MCTS1 | 10774 | -0.470 | -0.2004 | No | ||
| 90 | SLITRK6 | 10951 | -0.505 | -0.2075 | No | ||
| 91 | HOXD10 | 10979 | -0.513 | -0.2064 | No | ||
| 92 | NFATC4 | 11005 | -0.520 | -0.2051 | No | ||
| 93 | GJB1 | 11090 | -0.535 | -0.2070 | No | ||
| 94 | NEDD4 | 11123 | -0.540 | -0.2060 | No | ||
| 95 | HSPB2 | 11218 | -0.558 | -0.2084 | No | ||
| 96 | TYRO3 | 11576 | -0.627 | -0.2246 | No | ||
| 97 | KIRREL3 | 11850 | -0.681 | -0.2360 | No | ||
| 98 | GRIK3 | 11987 | -0.705 | -0.2398 | No | ||
| 99 | RARG | 11991 | -0.706 | -0.2365 | No | ||
| 100 | MYH11 | 12121 | -0.729 | -0.2398 | No | ||
| 101 | SALL2 | 12270 | -0.762 | -0.2440 | No | ||
| 102 | ATP5G2 | 12328 | -0.777 | -0.2432 | No | ||
| 103 | DHRS3 | 12347 | -0.781 | -0.2403 | No | ||
| 104 | SP3 | 12743 | -0.869 | -0.2574 | No | ||
| 105 | ZIC4 | 12811 | -0.882 | -0.2566 | No | ||
| 106 | LZTS2 | 12876 | -0.894 | -0.2556 | No | ||
| 107 | PRDM1 | 13010 | -0.924 | -0.2582 | No | ||
| 108 | NHLH2 | 13093 | -0.940 | -0.2580 | No | ||
| 109 | LOX | 13221 | -0.970 | -0.2600 | No | ||
| 110 | DNAJC5B | 13444 | -1.018 | -0.2670 | No | ||
| 111 | ZADH2 | 13784 | -1.094 | -0.2799 | No | ||
| 112 | GRM1 | 13805 | -1.103 | -0.2755 | No | ||
| 113 | FGF12 | 13816 | -1.104 | -0.2705 | No | ||
| 114 | GRIA3 | 14099 | -1.169 | -0.2800 | No | ||
| 115 | HIPK3 | 14349 | -1.232 | -0.2873 | Yes | ||
| 116 | PTCH2 | 14422 | -1.252 | -0.2850 | Yes | ||
| 117 | HOXA2 | 14503 | -1.270 | -0.2830 | Yes | ||
| 118 | EMX2 | 14515 | -1.273 | -0.2772 | Yes | ||
| 119 | BDNF | 14578 | -1.287 | -0.2742 | Yes | ||
| 120 | NCAM2 | 14655 | -1.303 | -0.2718 | Yes | ||
| 121 | RAB11FIP1 | 14778 | -1.332 | -0.2717 | Yes | ||
| 122 | SLC17A6 | 14788 | -1.335 | -0.2656 | Yes | ||
| 123 | BMPR2 | 14947 | -1.375 | -0.2673 | Yes | ||
| 124 | ANKRA2 | 15011 | -1.393 | -0.2637 | Yes | ||
| 125 | ONECUT2 | 15417 | -1.507 | -0.2782 | Yes | ||
| 126 | LUZP1 | 15424 | -1.508 | -0.2710 | Yes | ||
| 127 | FGF13 | 15436 | -1.513 | -0.2640 | Yes | ||
| 128 | HOXC6 | 15488 | -1.529 | -0.2592 | Yes | ||
| 129 | NEUROD6 | 15708 | -1.597 | -0.2631 | Yes | ||
| 130 | NR2F2 | 15883 | -1.657 | -0.2642 | Yes | ||
| 131 | MAP2K5 | 15964 | -1.686 | -0.2602 | Yes | ||
| 132 | FZD2 | 16003 | -1.697 | -0.2537 | Yes | ||
| 133 | LRP1 | 16305 | -1.801 | -0.2611 | Yes | ||
| 134 | CHCHD7 | 16322 | -1.807 | -0.2529 | Yes | ||
| 135 | BUB3 | 16391 | -1.828 | -0.2475 | Yes | ||
| 136 | SOX5 | 16414 | -1.837 | -0.2395 | Yes | ||
| 137 | CYP17A1 | 16475 | -1.862 | -0.2335 | Yes | ||
| 138 | RHOB | 16546 | -1.893 | -0.2278 | Yes | ||
| 139 | MRPS23 | 16676 | -1.952 | -0.2251 | Yes | ||
| 140 | SEPHS2 | 16684 | -1.957 | -0.2157 | Yes | ||
| 141 | HOXC4 | 16696 | -1.962 | -0.2065 | Yes | ||
| 142 | NR2F1 | 16761 | -1.987 | -0.2001 | Yes | ||
| 143 | CHRDL1 | 16857 | -2.036 | -0.1951 | Yes | ||
| 144 | CASK | 17004 | -2.109 | -0.1925 | Yes | ||
| 145 | EXT1 | 17046 | -2.132 | -0.1840 | Yes | ||
| 146 | VAMP3 | 17128 | -2.180 | -0.1776 | Yes | ||
| 147 | IPO4 | 17200 | -2.228 | -0.1703 | Yes | ||
| 148 | ITGA8 | 17218 | -2.239 | -0.1600 | Yes | ||
| 149 | POGZ | 17240 | -2.251 | -0.1500 | Yes | ||
| 150 | ARRDC3 | 17301 | -2.290 | -0.1418 | Yes | ||
| 151 | SLC13A1 | 17315 | -2.301 | -0.1310 | Yes | ||
| 152 | ZFX | 17376 | -2.347 | -0.1226 | Yes | ||
| 153 | MTUS1 | 17655 | -2.556 | -0.1249 | Yes | ||
| 154 | ST5 | 17764 | -2.648 | -0.1175 | Yes | ||
| 155 | TGFB2 | 17851 | -2.728 | -0.1086 | Yes | ||
| 156 | PRKAG1 | 17877 | -2.749 | -0.0962 | Yes | ||
| 157 | TNFRSF8 | 17889 | -2.759 | -0.0831 | Yes | ||
| 158 | ACACA | 18291 | -3.340 | -0.0881 | Yes | ||
| 159 | LDB2 | 18395 | -3.626 | -0.0756 | Yes | ||
| 160 | NR2F6 | 18482 | -3.977 | -0.0604 | Yes | ||
| 161 | RAB1A | 18499 | -4.108 | -0.0408 | Yes | ||
| 162 | CSNK1E | 18540 | -4.504 | -0.0205 | Yes | ||
| 163 | SERTAD4 | 18572 | -4.928 | 0.0024 | Yes |
