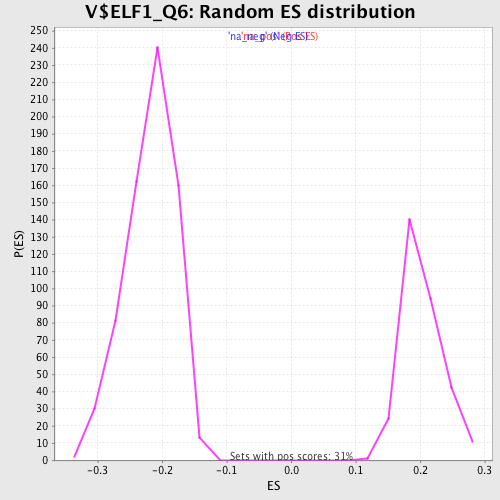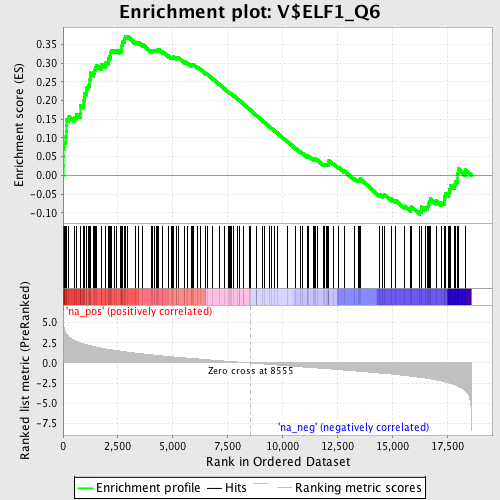
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$ELF1_Q6 |
| Enrichment Score (ES) | 0.3720006 |
| Normalized Enrichment Score (NES) | 1.8348771 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.059327137 |
| FWER p-Value | 0.141 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ATP2A3 | 6 | 5.724 | 0.0252 | Yes | ||
| 2 | VIM | 10 | 5.555 | 0.0498 | Yes | ||
| 3 | FGD2 | 24 | 4.704 | 0.0701 | Yes | ||
| 4 | ITGB7 | 41 | 4.403 | 0.0889 | Yes | ||
| 5 | MAP3K11 | 104 | 3.838 | 0.1027 | Yes | ||
| 6 | DAPP1 | 133 | 3.682 | 0.1176 | Yes | ||
| 7 | GMIP | 145 | 3.623 | 0.1332 | Yes | ||
| 8 | WAS | 159 | 3.582 | 0.1484 | Yes | ||
| 9 | PITPNM1 | 267 | 3.215 | 0.1570 | Yes | ||
| 10 | DMTF1 | 528 | 2.792 | 0.1553 | Yes | ||
| 11 | IQGAP1 | 617 | 2.684 | 0.1626 | Yes | ||
| 12 | ELMO1 | 786 | 2.490 | 0.1646 | Yes | ||
| 13 | DNAJA2 | 797 | 2.484 | 0.1751 | Yes | ||
| 14 | IKBKB | 798 | 2.484 | 0.1862 | Yes | ||
| 15 | CTCF | 918 | 2.376 | 0.1903 | Yes | ||
| 16 | GRB2 | 924 | 2.371 | 0.2007 | Yes | ||
| 17 | DGKZ | 965 | 2.341 | 0.2089 | Yes | ||
| 18 | APOB48R | 991 | 2.326 | 0.2180 | Yes | ||
| 19 | HM13 | 1053 | 2.286 | 0.2249 | Yes | ||
| 20 | CREBBP | 1062 | 2.275 | 0.2346 | Yes | ||
| 21 | MAP4K2 | 1136 | 2.202 | 0.2404 | Yes | ||
| 22 | ARPC2 | 1217 | 2.141 | 0.2457 | Yes | ||
| 23 | ABI3 | 1220 | 2.138 | 0.2551 | Yes | ||
| 24 | TTC15 | 1227 | 2.135 | 0.2643 | Yes | ||
| 25 | KLF13 | 1232 | 2.131 | 0.2736 | Yes | ||
| 26 | ADD3 | 1384 | 2.029 | 0.2745 | Yes | ||
| 27 | ELF4 | 1419 | 2.012 | 0.2816 | Yes | ||
| 28 | TLN1 | 1462 | 1.992 | 0.2882 | Yes | ||
| 29 | AIF1 | 1531 | 1.951 | 0.2932 | Yes | ||
| 30 | FBLN5 | 1732 | 1.839 | 0.2906 | Yes | ||
| 31 | PLCB2 | 1756 | 1.826 | 0.2975 | Yes | ||
| 32 | CDC14A | 1936 | 1.740 | 0.2956 | Yes | ||
| 33 | CD37 | 1951 | 1.731 | 0.3025 | Yes | ||
| 34 | MYADM | 2048 | 1.693 | 0.3049 | Yes | ||
| 35 | VAV1 | 2063 | 1.688 | 0.3117 | Yes | ||
| 36 | PUM1 | 2103 | 1.671 | 0.3170 | Yes | ||
| 37 | MYO1C | 2174 | 1.640 | 0.3205 | Yes | ||
| 38 | CAMK1D | 2178 | 1.638 | 0.3277 | Yes | ||
| 39 | A2BP1 | 2193 | 1.632 | 0.3342 | Yes | ||
| 40 | NASP | 2327 | 1.573 | 0.3340 | Yes | ||
| 41 | PCDH9 | 2448 | 1.522 | 0.3343 | Yes | ||
| 42 | PCQAP | 2592 | 1.469 | 0.3331 | Yes | ||
| 43 | STAT6 | 2672 | 1.439 | 0.3352 | Yes | ||
| 44 | NCAM1 | 2678 | 1.435 | 0.3414 | Yes | ||
| 45 | E2F3 | 2680 | 1.433 | 0.3477 | Yes | ||
| 46 | TLR4 | 2695 | 1.430 | 0.3533 | Yes | ||
| 47 | SORL1 | 2718 | 1.423 | 0.3585 | Yes | ||
| 48 | RNF31 | 2783 | 1.396 | 0.3612 | Yes | ||
| 49 | RUFY1 | 2801 | 1.388 | 0.3665 | Yes | ||
| 50 | LAG3 | 2822 | 1.380 | 0.3716 | Yes | ||
| 51 | PPP3CA | 2926 | 1.342 | 0.3720 | Yes | ||
| 52 | SAMSN1 | 3317 | 1.215 | 0.3563 | No | ||
| 53 | P2RY10 | 3421 | 1.185 | 0.3560 | No | ||
| 54 | RELA | 3628 | 1.116 | 0.3498 | No | ||
| 55 | RPL17 | 4034 | 0.994 | 0.3323 | No | ||
| 56 | ARHGDIB | 4075 | 0.983 | 0.3345 | No | ||
| 57 | RNF12 | 4151 | 0.962 | 0.3348 | No | ||
| 58 | GPX1 | 4238 | 0.938 | 0.3343 | No | ||
| 59 | BIRC4 | 4299 | 0.926 | 0.3352 | No | ||
| 60 | MLLT3 | 4362 | 0.909 | 0.3359 | No | ||
| 61 | SNX2 | 4547 | 0.857 | 0.3297 | No | ||
| 62 | NCK1 | 4808 | 0.793 | 0.3192 | No | ||
| 63 | ARRB2 | 4939 | 0.761 | 0.3155 | No | ||
| 64 | FCER1G | 5006 | 0.746 | 0.3153 | No | ||
| 65 | ALOX5AP | 5043 | 0.738 | 0.3166 | No | ||
| 66 | FUT11 | 5186 | 0.705 | 0.3121 | No | ||
| 67 | PI16 | 5188 | 0.705 | 0.3152 | No | ||
| 68 | SLC9A9 | 5236 | 0.693 | 0.3157 | No | ||
| 69 | FAM3D | 5541 | 0.624 | 0.3020 | No | ||
| 70 | SHC3 | 5546 | 0.622 | 0.3046 | No | ||
| 71 | TXK | 5686 | 0.595 | 0.2997 | No | ||
| 72 | TNFSF13 | 5861 | 0.556 | 0.2928 | No | ||
| 73 | PIK3CD | 5865 | 0.555 | 0.2951 | No | ||
| 74 | RAB39 | 5905 | 0.547 | 0.2954 | No | ||
| 75 | EDG6 | 5927 | 0.542 | 0.2967 | No | ||
| 76 | ORMDL3 | 6139 | 0.493 | 0.2875 | No | ||
| 77 | LIMS1 | 6247 | 0.471 | 0.2838 | No | ||
| 78 | TM6SF1 | 6510 | 0.411 | 0.2714 | No | ||
| 79 | XPR1 | 6560 | 0.399 | 0.2705 | No | ||
| 80 | KLF12 | 6826 | 0.342 | 0.2577 | No | ||
| 81 | FGF20 | 7125 | 0.277 | 0.2428 | No | ||
| 82 | MTPN | 7332 | 0.236 | 0.2327 | No | ||
| 83 | FZD10 | 7539 | 0.193 | 0.2224 | No | ||
| 84 | TYROBP | 7568 | 0.189 | 0.2217 | No | ||
| 85 | REL | 7630 | 0.179 | 0.2192 | No | ||
| 86 | ITPR2 | 7687 | 0.169 | 0.2169 | No | ||
| 87 | DPP8 | 7785 | 0.151 | 0.2123 | No | ||
| 88 | SIX4 | 7947 | 0.116 | 0.2041 | No | ||
| 89 | ESM1 | 8034 | 0.103 | 0.1999 | No | ||
| 90 | NR2C2 | 8036 | 0.103 | 0.2003 | No | ||
| 91 | CSF1 | 8235 | 0.068 | 0.1899 | No | ||
| 92 | RASGRP3 | 8241 | 0.067 | 0.1899 | No | ||
| 93 | TWIST1 | 8516 | 0.006 | 0.1751 | No | ||
| 94 | AKT1S1 | 8529 | 0.004 | 0.1745 | No | ||
| 95 | JARID2 | 8796 | -0.048 | 0.1603 | No | ||
| 96 | TGFB1 | 8803 | -0.050 | 0.1602 | No | ||
| 97 | CAMK2B | 8806 | -0.050 | 0.1603 | No | ||
| 98 | MR1 | 9077 | -0.108 | 0.1462 | No | ||
| 99 | MITF | 9171 | -0.128 | 0.1417 | No | ||
| 100 | SUPT16H | 9419 | -0.182 | 0.1291 | No | ||
| 101 | CLIC1 | 9511 | -0.201 | 0.1251 | No | ||
| 102 | STAB1 | 9634 | -0.227 | 0.1195 | No | ||
| 103 | SPRED2 | 9760 | -0.258 | 0.1139 | No | ||
| 104 | HOXB4 | 10212 | -0.353 | 0.0910 | No | ||
| 105 | TBC1D8 | 10603 | -0.434 | 0.0718 | No | ||
| 106 | TEK | 10803 | -0.475 | 0.0632 | No | ||
| 107 | PRDM5 | 10913 | -0.498 | 0.0595 | No | ||
| 108 | NEDD4 | 11123 | -0.540 | 0.0506 | No | ||
| 109 | CNOT6L | 11132 | -0.541 | 0.0525 | No | ||
| 110 | INPP5F | 11197 | -0.553 | 0.0515 | No | ||
| 111 | CD79B | 11408 | -0.595 | 0.0428 | No | ||
| 112 | PAX6 | 11434 | -0.603 | 0.0442 | No | ||
| 113 | CDKN2A | 11488 | -0.612 | 0.0440 | No | ||
| 114 | ELOVL1 | 11589 | -0.629 | 0.0414 | No | ||
| 115 | PSPN | 11880 | -0.686 | 0.0288 | No | ||
| 116 | C1QA | 11919 | -0.693 | 0.0298 | No | ||
| 117 | PTPRH | 12000 | -0.706 | 0.0286 | No | ||
| 118 | ERF | 12033 | -0.712 | 0.0300 | No | ||
| 119 | TSSC1 | 12098 | -0.724 | 0.0298 | No | ||
| 120 | CALCRL | 12100 | -0.725 | 0.0330 | No | ||
| 121 | RAC1 | 12108 | -0.727 | 0.0359 | No | ||
| 122 | MYL1 | 12114 | -0.728 | 0.0388 | No | ||
| 123 | MLLT7 | 12316 | -0.775 | 0.0314 | No | ||
| 124 | MRPL24 | 12568 | -0.828 | 0.0215 | No | ||
| 125 | NFIA | 12805 | -0.880 | 0.0126 | No | ||
| 126 | DPP3 | 13276 | -0.981 | -0.0084 | No | ||
| 127 | ACSL4 | 13440 | -1.018 | -0.0127 | No | ||
| 128 | IL7R | 13502 | -1.030 | -0.0114 | No | ||
| 129 | RASIP1 | 13540 | -1.039 | -0.0088 | No | ||
| 130 | KIF3C | 14436 | -1.255 | -0.0517 | No | ||
| 131 | AGPAT1 | 14572 | -1.286 | -0.0533 | No | ||
| 132 | MADD | 14631 | -1.298 | -0.0506 | No | ||
| 133 | STIM1 | 14966 | -1.383 | -0.0626 | No | ||
| 134 | CDH5 | 15164 | -1.429 | -0.0669 | No | ||
| 135 | CAPN3 | 15568 | -1.553 | -0.0818 | No | ||
| 136 | MSH5 | 15834 | -1.639 | -0.0888 | No | ||
| 137 | MARCKS | 15872 | -1.651 | -0.0834 | No | ||
| 138 | LAMA3 | 16261 | -1.784 | -0.0965 | No | ||
| 139 | CMAS | 16320 | -1.806 | -0.0916 | No | ||
| 140 | COL6A3 | 16334 | -1.811 | -0.0842 | No | ||
| 141 | GNGT2 | 16534 | -1.887 | -0.0866 | No | ||
| 142 | HOXC10 | 16629 | -1.929 | -0.0831 | No | ||
| 143 | EIF4EBP1 | 16641 | -1.937 | -0.0750 | No | ||
| 144 | HOXC4 | 16696 | -1.962 | -0.0692 | No | ||
| 145 | GPR120 | 16721 | -1.972 | -0.0617 | No | ||
| 146 | RAB25 | 16996 | -2.105 | -0.0671 | No | ||
| 147 | DUSP22 | 17261 | -2.266 | -0.0713 | No | ||
| 148 | CENTA1 | 17366 | -2.340 | -0.0665 | No | ||
| 149 | KCNAB1 | 17396 | -2.365 | -0.0575 | No | ||
| 150 | RALA | 17413 | -2.375 | -0.0478 | No | ||
| 151 | RNF5 | 17583 | -2.486 | -0.0459 | No | ||
| 152 | IL2 | 17618 | -2.529 | -0.0364 | No | ||
| 153 | RCL1 | 17657 | -2.559 | -0.0271 | No | ||
| 154 | NTS | 17823 | -2.698 | -0.0240 | No | ||
| 155 | PSME2 | 17897 | -2.763 | -0.0156 | No | ||
| 156 | CREB3 | 17981 | -2.878 | -0.0072 | No | ||
| 157 | PABPC1 | 17983 | -2.882 | 0.0056 | No | ||
| 158 | PRKRIP1 | 18000 | -2.897 | 0.0176 | No | ||
| 159 | TBC1D17 | 18347 | -3.519 | 0.0146 | No |