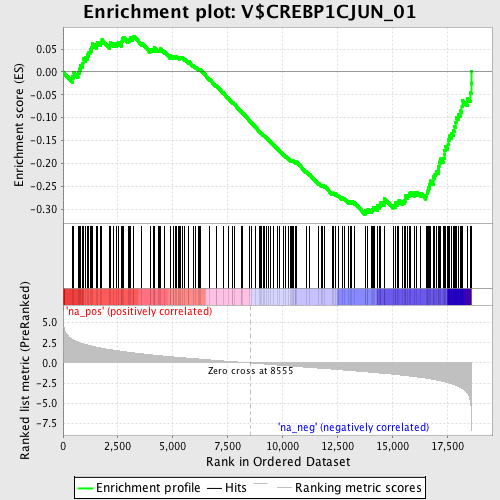
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$CREBP1CJUN_01 |
| Enrichment Score (ES) | -0.31196314 |
| Normalized Enrichment Score (NES) | -1.457124 |
| Nominal p-value | 0.00862069 |
| FDR q-value | 0.63368744 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SENP2 | 414 | 2.935 | -0.0099 | No | ||
| 2 | STAT3 | 479 | 2.857 | -0.0011 | No | ||
| 3 | SPAG9 | 681 | 2.608 | -0.0009 | No | ||
| 4 | SYNCRIP | 744 | 2.528 | 0.0065 | No | ||
| 5 | IKBKB | 798 | 2.484 | 0.0143 | No | ||
| 6 | BRE | 897 | 2.396 | 0.0192 | No | ||
| 7 | USP48 | 908 | 2.384 | 0.0289 | No | ||
| 8 | ZFYVE27 | 1034 | 2.300 | 0.0319 | No | ||
| 9 | RAI1 | 1109 | 2.230 | 0.0375 | No | ||
| 10 | UBE2H | 1175 | 2.174 | 0.0432 | No | ||
| 11 | GTF3C1 | 1228 | 2.135 | 0.0495 | No | ||
| 12 | EEF2 | 1310 | 2.074 | 0.0540 | No | ||
| 13 | ANAPC10 | 1336 | 2.055 | 0.0614 | No | ||
| 14 | PPP2R2A | 1523 | 1.957 | 0.0597 | No | ||
| 15 | SLC18A2 | 1575 | 1.924 | 0.0652 | No | ||
| 16 | RNF44 | 1722 | 1.842 | 0.0651 | No | ||
| 17 | RPS29 | 1771 | 1.822 | 0.0703 | No | ||
| 18 | MAFF | 2129 | 1.659 | 0.0581 | No | ||
| 19 | ARIH1 | 2137 | 1.656 | 0.0648 | No | ||
| 20 | TEX14 | 2295 | 1.586 | 0.0630 | No | ||
| 21 | DCTN1 | 2441 | 1.525 | 0.0617 | No | ||
| 22 | PNMA3 | 2511 | 1.502 | 0.0644 | No | ||
| 23 | SNF1LK | 2669 | 1.442 | 0.0620 | No | ||
| 24 | NCAM1 | 2678 | 1.435 | 0.0677 | No | ||
| 25 | THOC1 | 2689 | 1.431 | 0.0733 | No | ||
| 26 | CLCN3 | 2756 | 1.406 | 0.0757 | No | ||
| 27 | CBX8 | 2964 | 1.332 | 0.0702 | No | ||
| 28 | PHC1 | 3044 | 1.307 | 0.0715 | No | ||
| 29 | LENG9 | 3082 | 1.297 | 0.0751 | No | ||
| 30 | HOXB9 | 3185 | 1.260 | 0.0749 | No | ||
| 31 | CSTF3 | 3219 | 1.249 | 0.0785 | No | ||
| 32 | OSR1 | 3593 | 1.127 | 0.0631 | No | ||
| 33 | DDX51 | 3962 | 1.011 | 0.0474 | No | ||
| 34 | SUV39H2 | 3992 | 1.006 | 0.0502 | No | ||
| 35 | TNFAIP1 | 4098 | 0.977 | 0.0486 | No | ||
| 36 | ST13 | 4155 | 0.961 | 0.0497 | No | ||
| 37 | TRPC1 | 4163 | 0.958 | 0.0534 | No | ||
| 38 | PKP4 | 4343 | 0.916 | 0.0476 | No | ||
| 39 | PITX2 | 4409 | 0.896 | 0.0479 | No | ||
| 40 | SEC24C | 4428 | 0.890 | 0.0508 | No | ||
| 41 | CD2AP | 4612 | 0.843 | 0.0445 | No | ||
| 42 | NF1 | 4892 | 0.773 | 0.0326 | No | ||
| 43 | SCAMP5 | 4903 | 0.770 | 0.0354 | No | ||
| 44 | CRH | 5011 | 0.745 | 0.0328 | No | ||
| 45 | DNTTIP1 | 5046 | 0.737 | 0.0341 | No | ||
| 46 | SLC7A3 | 5102 | 0.723 | 0.0342 | No | ||
| 47 | PPM1A | 5179 | 0.707 | 0.0331 | No | ||
| 48 | IRX6 | 5260 | 0.686 | 0.0317 | No | ||
| 49 | PER1 | 5326 | 0.668 | 0.0310 | No | ||
| 50 | SP1 | 5357 | 0.661 | 0.0322 | No | ||
| 51 | ADCY8 | 5427 | 0.645 | 0.0312 | No | ||
| 52 | B3GALT2 | 5538 | 0.624 | 0.0279 | No | ||
| 53 | NDUFA10 | 5711 | 0.590 | 0.0211 | No | ||
| 54 | DHX36 | 5732 | 0.588 | 0.0226 | No | ||
| 55 | SNAP25 | 5924 | 0.543 | 0.0145 | No | ||
| 56 | AREG | 6035 | 0.517 | 0.0108 | No | ||
| 57 | SST | 6189 | 0.485 | 0.0046 | No | ||
| 58 | TRAP1 | 6228 | 0.475 | 0.0045 | No | ||
| 59 | CDX4 | 6249 | 0.471 | 0.0055 | No | ||
| 60 | MBNL2 | 6693 | 0.370 | -0.0170 | No | ||
| 61 | CHGB | 6970 | 0.309 | -0.0306 | No | ||
| 62 | CYLD | 6978 | 0.308 | -0.0297 | No | ||
| 63 | MRGPRF | 7290 | 0.244 | -0.0455 | No | ||
| 64 | RPL41 | 7515 | 0.197 | -0.0568 | No | ||
| 65 | CENPE | 7732 | 0.161 | -0.0678 | No | ||
| 66 | RAB6IP1 | 7734 | 0.160 | -0.0672 | No | ||
| 67 | HS3ST3A1 | 7821 | 0.143 | -0.0713 | No | ||
| 68 | SLC38A1 | 8116 | 0.088 | -0.0868 | No | ||
| 69 | AVPI1 | 8152 | 0.081 | -0.0884 | No | ||
| 70 | PHACTR3 | 8180 | 0.077 | -0.0895 | No | ||
| 71 | KCNA5 | 8515 | 0.007 | -0.1076 | No | ||
| 72 | PRR3 | 8603 | -0.012 | -0.1123 | No | ||
| 73 | TH | 8755 | -0.042 | -0.1203 | No | ||
| 74 | MLF2 | 8941 | -0.079 | -0.1300 | No | ||
| 75 | NUP98 | 8992 | -0.087 | -0.1323 | No | ||
| 76 | UCN | 9059 | -0.103 | -0.1354 | No | ||
| 77 | PTPRU | 9146 | -0.124 | -0.1396 | No | ||
| 78 | MITF | 9171 | -0.128 | -0.1403 | No | ||
| 79 | CCNA2 | 9253 | -0.149 | -0.1441 | No | ||
| 80 | ELL2 | 9371 | -0.171 | -0.1497 | No | ||
| 81 | PNRC1 | 9429 | -0.183 | -0.1520 | No | ||
| 82 | ELAVL1 | 9576 | -0.215 | -0.1590 | No | ||
| 83 | AHI1 | 9755 | -0.256 | -0.1676 | No | ||
| 84 | ATP6V0C | 9859 | -0.276 | -0.1720 | No | ||
| 85 | HHIP | 10062 | -0.324 | -0.1815 | No | ||
| 86 | ZBTB11 | 10156 | -0.340 | -0.1851 | No | ||
| 87 | PPARGC1A | 10259 | -0.366 | -0.1891 | No | ||
| 88 | MAOA | 10377 | -0.390 | -0.1938 | No | ||
| 89 | SEMA4C | 10410 | -0.396 | -0.1938 | No | ||
| 90 | SYNGR3 | 10468 | -0.407 | -0.1952 | No | ||
| 91 | OSBPL9 | 10493 | -0.411 | -0.1947 | No | ||
| 92 | OGDH | 10608 | -0.434 | -0.1990 | No | ||
| 93 | PCSK1 | 10609 | -0.434 | -0.1972 | No | ||
| 94 | CLSTN3 | 10639 | -0.440 | -0.1969 | No | ||
| 95 | CAMK2D | 11082 | -0.534 | -0.2185 | No | ||
| 96 | HAS1 | 11233 | -0.561 | -0.2243 | No | ||
| 97 | DIO2 | 11640 | -0.641 | -0.2436 | No | ||
| 98 | PAFAH1B1 | 11781 | -0.667 | -0.2483 | No | ||
| 99 | YTHDC2 | 11836 | -0.678 | -0.2483 | No | ||
| 100 | ETV6 | 11933 | -0.696 | -0.2506 | No | ||
| 101 | IRX4 | 12267 | -0.762 | -0.2654 | No | ||
| 102 | SCG2 | 12321 | -0.776 | -0.2649 | No | ||
| 103 | FOXD3 | 12420 | -0.794 | -0.2668 | No | ||
| 104 | EPHA2 | 12535 | -0.820 | -0.2695 | No | ||
| 105 | KCTD8 | 12719 | -0.864 | -0.2757 | No | ||
| 106 | CDS1 | 12802 | -0.879 | -0.2764 | No | ||
| 107 | NR2E1 | 13004 | -0.923 | -0.2834 | No | ||
| 108 | ELOVL5 | 13106 | -0.944 | -0.2848 | No | ||
| 109 | CXCL16 | 13147 | -0.951 | -0.2829 | No | ||
| 110 | STAG1 | 13268 | -0.980 | -0.2852 | No | ||
| 111 | ABCE1 | 13762 | -1.089 | -0.3073 | Yes | ||
| 112 | RELB | 13763 | -1.089 | -0.3026 | Yes | ||
| 113 | LMCD1 | 13872 | -1.118 | -0.3037 | Yes | ||
| 114 | SLC25A25 | 13893 | -1.123 | -0.3000 | Yes | ||
| 115 | GNB4 | 14053 | -1.159 | -0.3037 | Yes | ||
| 116 | NDUFB2 | 14095 | -1.168 | -0.3009 | Yes | ||
| 117 | SYT11 | 14127 | -1.177 | -0.2975 | Yes | ||
| 118 | RCE1 | 14187 | -1.192 | -0.2956 | Yes | ||
| 119 | BAT5 | 14318 | -1.224 | -0.2975 | Yes | ||
| 120 | PEG3 | 14328 | -1.227 | -0.2927 | Yes | ||
| 121 | MAPK10 | 14428 | -1.253 | -0.2927 | Yes | ||
| 122 | RAD51C | 14467 | -1.260 | -0.2894 | Yes | ||
| 123 | ING4 | 14486 | -1.266 | -0.2849 | Yes | ||
| 124 | MADD | 14631 | -1.298 | -0.2872 | Yes | ||
| 125 | FLT1 | 14634 | -1.298 | -0.2818 | Yes | ||
| 126 | LDHA | 14647 | -1.302 | -0.2768 | Yes | ||
| 127 | PPM2C | 15037 | -1.396 | -0.2920 | Yes | ||
| 128 | RBKS | 15136 | -1.422 | -0.2912 | Yes | ||
| 129 | YME1L1 | 15146 | -1.424 | -0.2856 | Yes | ||
| 130 | CLDN7 | 15259 | -1.453 | -0.2855 | Yes | ||
| 131 | PARD6A | 15307 | -1.469 | -0.2817 | Yes | ||
| 132 | HCRTR2 | 15469 | -1.524 | -0.2839 | Yes | ||
| 133 | THADA | 15539 | -1.544 | -0.2811 | Yes | ||
| 134 | CREM | 15586 | -1.558 | -0.2769 | Yes | ||
| 135 | RUSC1 | 15602 | -1.563 | -0.2710 | Yes | ||
| 136 | NEUROD6 | 15708 | -1.597 | -0.2699 | Yes | ||
| 137 | WISP1 | 15777 | -1.619 | -0.2667 | Yes | ||
| 138 | MCAM | 15838 | -1.641 | -0.2629 | Yes | ||
| 139 | RBM18 | 16007 | -1.698 | -0.2648 | Yes | ||
| 140 | NCALD | 16121 | -1.739 | -0.2634 | Yes | ||
| 141 | MYL6 | 16292 | -1.797 | -0.2650 | Yes | ||
| 142 | DSCR1 | 16539 | -1.890 | -0.2702 | Yes | ||
| 143 | ZHX2 | 16585 | -1.911 | -0.2645 | Yes | ||
| 144 | HOXC10 | 16629 | -1.929 | -0.2586 | Yes | ||
| 145 | WNT10A | 16669 | -1.949 | -0.2524 | Yes | ||
| 146 | SULT4A1 | 16707 | -1.966 | -0.2460 | Yes | ||
| 147 | NOL4 | 16726 | -1.973 | -0.2385 | Yes | ||
| 148 | DAAM2 | 16875 | -2.046 | -0.2378 | Yes | ||
| 149 | GEM | 16878 | -2.048 | -0.2291 | Yes | ||
| 150 | EGR1 | 16946 | -2.080 | -0.2239 | Yes | ||
| 151 | RAB25 | 16996 | -2.105 | -0.2175 | Yes | ||
| 152 | GPM6B | 17094 | -2.160 | -0.2136 | Yes | ||
| 153 | GPR3 | 17125 | -2.179 | -0.2059 | Yes | ||
| 154 | DUSP1 | 17135 | -2.182 | -0.1970 | Yes | ||
| 155 | UMPS | 17188 | -2.221 | -0.1903 | Yes | ||
| 156 | RNF7 | 17338 | -2.318 | -0.1885 | Yes | ||
| 157 | CENTA1 | 17366 | -2.340 | -0.1800 | Yes | ||
| 158 | MRRF | 17392 | -2.362 | -0.1712 | Yes | ||
| 159 | TRAF4 | 17416 | -2.376 | -0.1623 | Yes | ||
| 160 | MAP3K13 | 17541 | -2.451 | -0.1586 | Yes | ||
| 161 | LTBP1 | 17552 | -2.457 | -0.1486 | Yes | ||
| 162 | YWHAZ | 17591 | -2.498 | -0.1400 | Yes | ||
| 163 | MAP1LC3A | 17695 | -2.592 | -0.1345 | Yes | ||
| 164 | PLCD3 | 17799 | -2.678 | -0.1286 | Yes | ||
| 165 | ARL6IP2 | 17831 | -2.708 | -0.1187 | Yes | ||
| 166 | ALS2 | 17896 | -2.763 | -0.1104 | Yes | ||
| 167 | NUPL2 | 17912 | -2.783 | -0.0993 | Yes | ||
| 168 | PDLIM3 | 18018 | -2.920 | -0.0925 | Yes | ||
| 169 | TGIF2 | 18129 | -3.070 | -0.0853 | Yes | ||
| 170 | KCNF1 | 18179 | -3.147 | -0.0745 | Yes | ||
| 171 | CCBL1 | 18190 | -3.161 | -0.0616 | Yes | ||
| 172 | DDX28 | 18420 | -3.688 | -0.0582 | Yes | ||
| 173 | BNIP3L | 18558 | -4.653 | -0.0457 | Yes | ||
| 174 | NR4A2 | 18597 | -5.541 | -0.0241 | Yes | ||
| 175 | SDHB | 18604 | -5.865 | 0.0007 | Yes |
