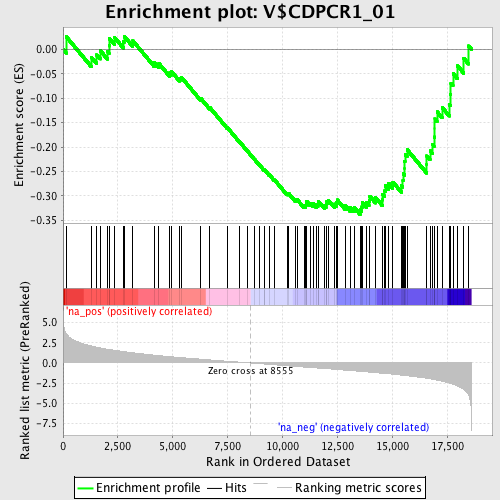
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$CDPCR1_01 |
| Enrichment Score (ES) | -0.33831653 |
| Normalized Enrichment Score (NES) | -1.4208223 |
| Nominal p-value | 0.036507938 |
| FDR q-value | 0.5167458 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GRB7 | 141 | 3.631 | 0.0263 | No | ||
| 2 | BHLHB3 | 1295 | 2.085 | -0.0164 | No | ||
| 3 | CIB3 | 1534 | 1.949 | -0.0111 | No | ||
| 4 | TLE4 | 1692 | 1.857 | -0.0022 | No | ||
| 5 | SERPING1 | 2021 | 1.703 | -0.0040 | No | ||
| 6 | PFN2 | 2097 | 1.676 | 0.0076 | No | ||
| 7 | CSMD3 | 2108 | 1.668 | 0.0226 | No | ||
| 8 | MBNL1 | 2349 | 1.564 | 0.0243 | No | ||
| 9 | SON | 2748 | 1.410 | 0.0160 | No | ||
| 10 | PAX8 | 2780 | 1.397 | 0.0274 | No | ||
| 11 | THRA | 3180 | 1.263 | 0.0176 | No | ||
| 12 | PLXNA3 | 4158 | 0.960 | -0.0261 | No | ||
| 13 | IL15 | 4365 | 0.909 | -0.0288 | No | ||
| 14 | KLF15 | 4858 | 0.781 | -0.0480 | No | ||
| 15 | TBX6 | 4943 | 0.761 | -0.0455 | No | ||
| 16 | AP3M2 | 5305 | 0.674 | -0.0586 | No | ||
| 17 | DGKB | 5381 | 0.655 | -0.0566 | No | ||
| 18 | CYP26B1 | 6264 | 0.468 | -0.0998 | No | ||
| 19 | EMP1 | 6684 | 0.371 | -0.1189 | No | ||
| 20 | NOG | 7484 | 0.203 | -0.1602 | No | ||
| 21 | TRIB2 | 8056 | 0.099 | -0.1900 | No | ||
| 22 | MEIS1 | 8414 | 0.030 | -0.2090 | No | ||
| 23 | VAX1 | 8736 | -0.038 | -0.2260 | No | ||
| 24 | MYST2 | 8952 | -0.081 | -0.2368 | No | ||
| 25 | STAG2 | 9164 | -0.127 | -0.2470 | No | ||
| 26 | LIN28 | 9180 | -0.130 | -0.2466 | No | ||
| 27 | NKX2-2 | 9397 | -0.177 | -0.2566 | No | ||
| 28 | ERG | 9635 | -0.228 | -0.2673 | No | ||
| 29 | HOXB4 | 10212 | -0.353 | -0.2951 | No | ||
| 30 | NR3C2 | 10269 | -0.369 | -0.2947 | No | ||
| 31 | NRXN3 | 10596 | -0.433 | -0.3082 | No | ||
| 32 | ADAM10 | 10666 | -0.446 | -0.3078 | No | ||
| 33 | HOXD10 | 10979 | -0.513 | -0.3198 | No | ||
| 34 | PIGN | 11062 | -0.530 | -0.3193 | No | ||
| 35 | INSM2 | 11078 | -0.534 | -0.3151 | No | ||
| 36 | LYPLA3 | 11103 | -0.537 | -0.3114 | No | ||
| 37 | COL2A1 | 11269 | -0.568 | -0.3150 | No | ||
| 38 | FCHSD1 | 11392 | -0.592 | -0.3161 | No | ||
| 39 | RPH3A | 11542 | -0.621 | -0.3183 | No | ||
| 40 | BRUNOL4 | 11619 | -0.636 | -0.3165 | No | ||
| 41 | DIO2 | 11640 | -0.641 | -0.3116 | No | ||
| 42 | MGAT5B | 11905 | -0.690 | -0.3194 | No | ||
| 43 | DBC1 | 11986 | -0.705 | -0.3171 | No | ||
| 44 | SFTPC | 12020 | -0.709 | -0.3122 | No | ||
| 45 | TCF7L2 | 12099 | -0.725 | -0.3097 | No | ||
| 46 | STMN2 | 12368 | -0.785 | -0.3168 | No | ||
| 47 | GATA4 | 12454 | -0.801 | -0.3139 | No | ||
| 48 | SLC12A5 | 12492 | -0.809 | -0.3084 | No | ||
| 49 | ETV1 | 12869 | -0.893 | -0.3203 | No | ||
| 50 | NHLH2 | 13093 | -0.940 | -0.3236 | No | ||
| 51 | STAT5B | 13272 | -0.980 | -0.3240 | No | ||
| 52 | PDCD10 | 13538 | -1.039 | -0.3286 | Yes | ||
| 53 | NTRK1 | 13616 | -1.059 | -0.3229 | Yes | ||
| 54 | TBX3 | 13629 | -1.061 | -0.3136 | Yes | ||
| 55 | FGF12 | 13816 | -1.104 | -0.3133 | Yes | ||
| 56 | DPP10 | 13945 | -1.136 | -0.3096 | Yes | ||
| 57 | GART | 13979 | -1.143 | -0.3007 | Yes | ||
| 58 | DUSP4 | 14241 | -1.206 | -0.3036 | Yes | ||
| 59 | MAG | 14552 | -1.282 | -0.3083 | Yes | ||
| 60 | MAOB | 14560 | -1.283 | -0.2967 | Yes | ||
| 61 | ITGA7 | 14662 | -1.305 | -0.2900 | Yes | ||
| 62 | RTN4RL2 | 14690 | -1.311 | -0.2792 | Yes | ||
| 63 | BTBD3 | 14835 | -1.345 | -0.2744 | Yes | ||
| 64 | EEF1A2 | 15028 | -1.396 | -0.2717 | Yes | ||
| 65 | NFIB | 15419 | -1.508 | -0.2787 | Yes | ||
| 66 | HOXC6 | 15488 | -1.529 | -0.2681 | Yes | ||
| 67 | RFX4 | 15500 | -1.533 | -0.2543 | Yes | ||
| 68 | CPNE6 | 15566 | -1.551 | -0.2434 | Yes | ||
| 69 | KCND1 | 15570 | -1.554 | -0.2290 | Yes | ||
| 70 | CREM | 15586 | -1.558 | -0.2153 | Yes | ||
| 71 | GRIK4 | 15679 | -1.589 | -0.2054 | Yes | ||
| 72 | PPT2 | 16573 | -1.903 | -0.2358 | Yes | ||
| 73 | HOXA10 | 16579 | -1.907 | -0.2183 | Yes | ||
| 74 | NOL4 | 16726 | -1.973 | -0.2077 | Yes | ||
| 75 | NTNG2 | 16830 | -2.023 | -0.1944 | Yes | ||
| 76 | INSRR | 16910 | -2.061 | -0.1794 | Yes | ||
| 77 | EGR1 | 16946 | -2.080 | -0.1619 | Yes | ||
| 78 | HOXC5 | 16947 | -2.080 | -0.1424 | Yes | ||
| 79 | EXT1 | 17046 | -2.132 | -0.1278 | Yes | ||
| 80 | ST7 | 17276 | -2.274 | -0.1189 | Yes | ||
| 81 | HOXB2 | 17604 | -2.511 | -0.1131 | Yes | ||
| 82 | KCNS1 | 17654 | -2.556 | -0.0919 | Yes | ||
| 83 | KDELR2 | 17675 | -2.575 | -0.0689 | Yes | ||
| 84 | KCNA1 | 17773 | -2.652 | -0.0494 | Yes | ||
| 85 | SERPINI1 | 17977 | -2.877 | -0.0335 | Yes | ||
| 86 | CUL2 | 18261 | -3.287 | -0.0180 | Yes | ||
| 87 | NR2F6 | 18482 | -3.977 | 0.0072 | Yes |
