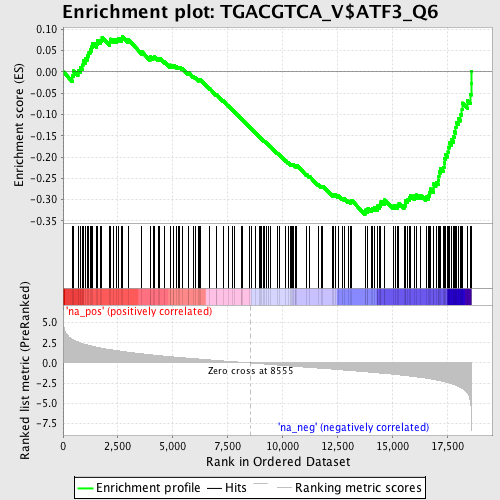
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TGACGTCA_V$ATF3_Q6 |
| Enrichment Score (ES) | -0.33474654 |
| Normalized Enrichment Score (NES) | -1.5291524 |
| Nominal p-value | 0.0014450867 |
| FDR q-value | 0.43884134 |
| FWER p-Value | 0.982 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SENP2 | 414 | 2.935 | -0.0082 | No | ||
| 2 | STAT3 | 479 | 2.857 | 0.0023 | No | ||
| 3 | SPAG9 | 681 | 2.608 | 0.0040 | No | ||
| 4 | IKBKB | 798 | 2.484 | 0.0098 | No | ||
| 5 | BRE | 897 | 2.396 | 0.0162 | No | ||
| 6 | USP48 | 908 | 2.384 | 0.0272 | No | ||
| 7 | ZFYVE27 | 1034 | 2.300 | 0.0316 | No | ||
| 8 | RAI1 | 1109 | 2.230 | 0.0384 | No | ||
| 9 | UBE2H | 1175 | 2.174 | 0.0455 | No | ||
| 10 | GTF3C1 | 1228 | 2.135 | 0.0530 | No | ||
| 11 | EEF2 | 1310 | 2.074 | 0.0587 | No | ||
| 12 | ANAPC10 | 1336 | 2.055 | 0.0674 | No | ||
| 13 | PPP2R2A | 1523 | 1.957 | 0.0668 | No | ||
| 14 | SLC18A2 | 1575 | 1.924 | 0.0734 | No | ||
| 15 | RNF44 | 1722 | 1.842 | 0.0744 | No | ||
| 16 | RPS29 | 1771 | 1.822 | 0.0807 | No | ||
| 17 | MAFF | 2129 | 1.659 | 0.0694 | No | ||
| 18 | ARIH1 | 2137 | 1.656 | 0.0771 | No | ||
| 19 | TEX14 | 2295 | 1.586 | 0.0763 | No | ||
| 20 | DCTN1 | 2441 | 1.525 | 0.0758 | No | ||
| 21 | PNMA3 | 2511 | 1.502 | 0.0794 | No | ||
| 22 | SNF1LK | 2669 | 1.442 | 0.0779 | No | ||
| 23 | THOC1 | 2689 | 1.431 | 0.0838 | No | ||
| 24 | CBX8 | 2964 | 1.332 | 0.0755 | No | ||
| 25 | OSR1 | 3593 | 1.127 | 0.0469 | No | ||
| 26 | DDX51 | 3962 | 1.011 | 0.0319 | No | ||
| 27 | SUV39H2 | 3992 | 1.006 | 0.0352 | No | ||
| 28 | TNFAIP1 | 4098 | 0.977 | 0.0343 | No | ||
| 29 | ST13 | 4155 | 0.961 | 0.0359 | No | ||
| 30 | PKP4 | 4343 | 0.916 | 0.0302 | No | ||
| 31 | PITX2 | 4409 | 0.896 | 0.0311 | No | ||
| 32 | CD2AP | 4612 | 0.843 | 0.0242 | No | ||
| 33 | NF1 | 4892 | 0.773 | 0.0129 | No | ||
| 34 | SCAMP5 | 4903 | 0.770 | 0.0161 | No | ||
| 35 | CRH | 5011 | 0.745 | 0.0139 | No | ||
| 36 | DNTTIP1 | 5046 | 0.737 | 0.0157 | No | ||
| 37 | PPM1A | 5179 | 0.707 | 0.0119 | No | ||
| 38 | IRX6 | 5260 | 0.686 | 0.0109 | No | ||
| 39 | PER1 | 5326 | 0.668 | 0.0107 | No | ||
| 40 | ADCY8 | 5427 | 0.645 | 0.0084 | No | ||
| 41 | NDUFA10 | 5711 | 0.590 | -0.0041 | No | ||
| 42 | DHX36 | 5732 | 0.588 | -0.0023 | No | ||
| 43 | SNAP25 | 5924 | 0.543 | -0.0100 | No | ||
| 44 | AREG | 6035 | 0.517 | -0.0135 | No | ||
| 45 | SST | 6189 | 0.485 | -0.0194 | No | ||
| 46 | TRAP1 | 6228 | 0.475 | -0.0191 | No | ||
| 47 | CDX4 | 6249 | 0.471 | -0.0179 | No | ||
| 48 | MBNL2 | 6693 | 0.370 | -0.0401 | No | ||
| 49 | CHGB | 6970 | 0.309 | -0.0536 | No | ||
| 50 | CYLD | 6978 | 0.308 | -0.0524 | No | ||
| 51 | MRGPRF | 7290 | 0.244 | -0.0681 | No | ||
| 52 | RPL41 | 7515 | 0.197 | -0.0793 | No | ||
| 53 | CENPE | 7732 | 0.161 | -0.0902 | No | ||
| 54 | HS3ST3A1 | 7821 | 0.143 | -0.0943 | No | ||
| 55 | SLC38A1 | 8116 | 0.088 | -0.1098 | No | ||
| 56 | AVPI1 | 8152 | 0.081 | -0.1113 | No | ||
| 57 | PHACTR3 | 8180 | 0.077 | -0.1123 | No | ||
| 58 | KCNA5 | 8515 | 0.007 | -0.1304 | No | ||
| 59 | PRR3 | 8603 | -0.012 | -0.1351 | No | ||
| 60 | TH | 8755 | -0.042 | -0.1430 | No | ||
| 61 | MLF2 | 8941 | -0.079 | -0.1527 | No | ||
| 62 | NUP98 | 8992 | -0.087 | -0.1549 | No | ||
| 63 | UCN | 9059 | -0.103 | -0.1580 | No | ||
| 64 | PTPRU | 9146 | -0.124 | -0.1621 | No | ||
| 65 | MITF | 9171 | -0.128 | -0.1628 | No | ||
| 66 | CCNA2 | 9253 | -0.149 | -0.1664 | No | ||
| 67 | ELL2 | 9371 | -0.171 | -0.1719 | No | ||
| 68 | PNRC1 | 9429 | -0.183 | -0.1741 | No | ||
| 69 | AHI1 | 9755 | -0.256 | -0.1905 | No | ||
| 70 | ATP6V0C | 9859 | -0.276 | -0.1947 | No | ||
| 71 | ZBTB11 | 10156 | -0.340 | -0.2091 | No | ||
| 72 | PPARGC1A | 10259 | -0.366 | -0.2128 | No | ||
| 73 | MAOA | 10377 | -0.390 | -0.2173 | No | ||
| 74 | SEMA4C | 10410 | -0.396 | -0.2171 | No | ||
| 75 | SYNGR3 | 10468 | -0.407 | -0.2182 | No | ||
| 76 | OSBPL9 | 10493 | -0.411 | -0.2175 | No | ||
| 77 | OGDH | 10608 | -0.434 | -0.2216 | No | ||
| 78 | PCSK1 | 10609 | -0.434 | -0.2195 | No | ||
| 79 | CLSTN3 | 10639 | -0.440 | -0.2189 | No | ||
| 80 | CAMK2D | 11082 | -0.534 | -0.2402 | No | ||
| 81 | HAS1 | 11233 | -0.561 | -0.2456 | No | ||
| 82 | DIO2 | 11640 | -0.641 | -0.2645 | No | ||
| 83 | PAFAH1B1 | 11781 | -0.667 | -0.2688 | No | ||
| 84 | YTHDC2 | 11836 | -0.678 | -0.2685 | No | ||
| 85 | IRX4 | 12267 | -0.762 | -0.2881 | No | ||
| 86 | SCG2 | 12321 | -0.776 | -0.2872 | No | ||
| 87 | FOXD3 | 12420 | -0.794 | -0.2886 | No | ||
| 88 | EPHA2 | 12535 | -0.820 | -0.2908 | No | ||
| 89 | KCTD8 | 12719 | -0.864 | -0.2965 | No | ||
| 90 | CDS1 | 12802 | -0.879 | -0.2967 | No | ||
| 91 | NR2E1 | 13004 | -0.923 | -0.3031 | No | ||
| 92 | ELOVL5 | 13106 | -0.944 | -0.3040 | No | ||
| 93 | CXCL16 | 13147 | -0.951 | -0.3015 | No | ||
| 94 | ABCE1 | 13762 | -1.089 | -0.3295 | Yes | ||
| 95 | RELB | 13763 | -1.089 | -0.3242 | Yes | ||
| 96 | LMCD1 | 13872 | -1.118 | -0.3246 | Yes | ||
| 97 | SLC25A25 | 13893 | -1.123 | -0.3202 | Yes | ||
| 98 | GNB4 | 14053 | -1.159 | -0.3232 | Yes | ||
| 99 | NDUFB2 | 14095 | -1.168 | -0.3197 | Yes | ||
| 100 | RCE1 | 14187 | -1.192 | -0.3189 | Yes | ||
| 101 | BAT5 | 14318 | -1.224 | -0.3200 | Yes | ||
| 102 | PEG3 | 14328 | -1.227 | -0.3145 | Yes | ||
| 103 | MAPK10 | 14428 | -1.253 | -0.3137 | Yes | ||
| 104 | RAD51C | 14467 | -1.260 | -0.3097 | Yes | ||
| 105 | ING4 | 14486 | -1.266 | -0.3045 | Yes | ||
| 106 | FLT1 | 14634 | -1.298 | -0.3062 | Yes | ||
| 107 | LDHA | 14647 | -1.302 | -0.3005 | Yes | ||
| 108 | PPM2C | 15037 | -1.396 | -0.3148 | Yes | ||
| 109 | RBKS | 15136 | -1.422 | -0.3132 | Yes | ||
| 110 | CLDN7 | 15259 | -1.453 | -0.3127 | Yes | ||
| 111 | PARD6A | 15307 | -1.469 | -0.3081 | Yes | ||
| 112 | THADA | 15539 | -1.544 | -0.3131 | Yes | ||
| 113 | CREM | 15586 | -1.558 | -0.3080 | Yes | ||
| 114 | RUSC1 | 15602 | -1.563 | -0.3013 | Yes | ||
| 115 | NEUROD6 | 15708 | -1.597 | -0.2992 | Yes | ||
| 116 | WISP1 | 15777 | -1.619 | -0.2950 | Yes | ||
| 117 | MCAM | 15838 | -1.641 | -0.2903 | Yes | ||
| 118 | RBM18 | 16007 | -1.698 | -0.2911 | Yes | ||
| 119 | NCALD | 16121 | -1.739 | -0.2888 | Yes | ||
| 120 | MYL6 | 16292 | -1.797 | -0.2893 | Yes | ||
| 121 | DSCR1 | 16539 | -1.890 | -0.2934 | Yes | ||
| 122 | WNT10A | 16669 | -1.949 | -0.2909 | Yes | ||
| 123 | SULT4A1 | 16707 | -1.966 | -0.2834 | Yes | ||
| 124 | NOL4 | 16726 | -1.973 | -0.2748 | Yes | ||
| 125 | DAAM2 | 16875 | -2.046 | -0.2728 | Yes | ||
| 126 | GEM | 16878 | -2.048 | -0.2630 | Yes | ||
| 127 | RAB25 | 16996 | -2.105 | -0.2591 | Yes | ||
| 128 | GPM6B | 17094 | -2.160 | -0.2539 | Yes | ||
| 129 | GPR3 | 17125 | -2.179 | -0.2449 | Yes | ||
| 130 | DUSP1 | 17135 | -2.182 | -0.2348 | Yes | ||
| 131 | UMPS | 17188 | -2.221 | -0.2268 | Yes | ||
| 132 | RNF7 | 17338 | -2.318 | -0.2236 | Yes | ||
| 133 | CENTA1 | 17366 | -2.340 | -0.2137 | Yes | ||
| 134 | MRRF | 17392 | -2.362 | -0.2036 | Yes | ||
| 135 | TRAF4 | 17416 | -2.376 | -0.1933 | Yes | ||
| 136 | MAP3K13 | 17541 | -2.451 | -0.1881 | Yes | ||
| 137 | LTBP1 | 17552 | -2.457 | -0.1767 | Yes | ||
| 138 | YWHAZ | 17591 | -2.498 | -0.1666 | Yes | ||
| 139 | MAP1LC3A | 17695 | -2.592 | -0.1596 | Yes | ||
| 140 | PLCD3 | 17799 | -2.678 | -0.1522 | Yes | ||
| 141 | ARL6IP2 | 17831 | -2.708 | -0.1407 | Yes | ||
| 142 | ALS2 | 17896 | -2.763 | -0.1307 | Yes | ||
| 143 | NUPL2 | 17912 | -2.783 | -0.1180 | Yes | ||
| 144 | PDLIM3 | 18018 | -2.920 | -0.1095 | Yes | ||
| 145 | TGIF2 | 18129 | -3.070 | -0.1005 | Yes | ||
| 146 | KCNF1 | 18179 | -3.147 | -0.0879 | Yes | ||
| 147 | CCBL1 | 18190 | -3.161 | -0.0731 | Yes | ||
| 148 | DDX28 | 18420 | -3.688 | -0.0676 | Yes | ||
| 149 | BNIP3L | 18558 | -4.653 | -0.0524 | Yes | ||
| 150 | NR4A2 | 18597 | -5.541 | -0.0275 | Yes | ||
| 151 | SDHB | 18604 | -5.865 | 0.0006 | Yes |
