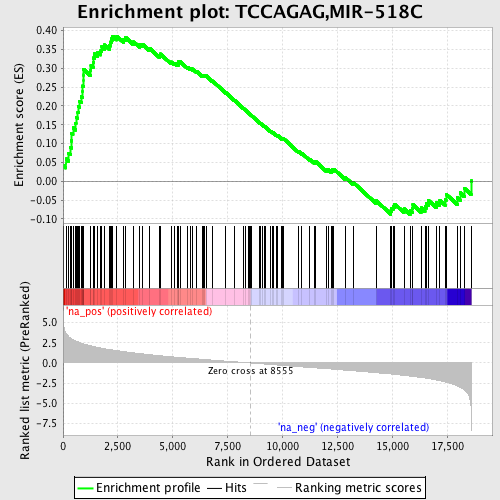
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TCCAGAG,MIR-518C |
| Enrichment Score (ES) | 0.38551354 |
| Normalized Enrichment Score (NES) | 1.8252168 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.05488355 |
| FWER p-Value | 0.155 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TCERG1 | 0 | 6.442 | 0.0423 | Yes | ||
| 2 | GAD1 | 131 | 3.695 | 0.0595 | Yes | ||
| 3 | RANBP10 | 256 | 3.241 | 0.0741 | Yes | ||
| 4 | CBX7 | 334 | 3.077 | 0.0901 | Yes | ||
| 5 | SLC38A2 | 374 | 2.993 | 0.1077 | Yes | ||
| 6 | MECP2 | 392 | 2.971 | 0.1262 | Yes | ||
| 7 | NFAT5 | 451 | 2.890 | 0.1421 | Yes | ||
| 8 | CDC42 | 567 | 2.734 | 0.1538 | Yes | ||
| 9 | NCF4 | 590 | 2.705 | 0.1704 | Yes | ||
| 10 | SMURF2 | 672 | 2.612 | 0.1831 | Yes | ||
| 11 | CSNK1G1 | 697 | 2.584 | 0.1988 | Yes | ||
| 12 | GGA3 | 755 | 2.513 | 0.2122 | Yes | ||
| 13 | PARP6 | 828 | 2.456 | 0.2245 | Yes | ||
| 14 | CAMK2G | 879 | 2.414 | 0.2376 | Yes | ||
| 15 | IL18BP | 885 | 2.408 | 0.2531 | Yes | ||
| 16 | TNK2 | 921 | 2.374 | 0.2668 | Yes | ||
| 17 | GRB2 | 924 | 2.371 | 0.2823 | Yes | ||
| 18 | LPHN1 | 928 | 2.368 | 0.2977 | Yes | ||
| 19 | NCOR2 | 1246 | 2.123 | 0.2945 | Yes | ||
| 20 | RBBP5 | 1269 | 2.106 | 0.3071 | Yes | ||
| 21 | ATP2A2 | 1370 | 2.038 | 0.3151 | Yes | ||
| 22 | ZDHHC9 | 1385 | 2.028 | 0.3276 | Yes | ||
| 23 | NFIX | 1429 | 2.005 | 0.3385 | Yes | ||
| 24 | PCDHGB1 | 1581 | 1.920 | 0.3429 | Yes | ||
| 25 | RNF44 | 1722 | 1.842 | 0.3474 | Yes | ||
| 26 | CHKB | 1765 | 1.824 | 0.3571 | Yes | ||
| 27 | SMPD3 | 1886 | 1.766 | 0.3622 | Yes | ||
| 28 | PUM1 | 2103 | 1.671 | 0.3615 | Yes | ||
| 29 | AIRE | 2149 | 1.650 | 0.3699 | Yes | ||
| 30 | A2BP1 | 2193 | 1.632 | 0.3783 | Yes | ||
| 31 | PHF21A | 2256 | 1.605 | 0.3855 | Yes | ||
| 32 | PCDH9 | 2448 | 1.522 | 0.3852 | No | ||
| 33 | GCLC | 2758 | 1.406 | 0.3777 | No | ||
| 34 | TAF4 | 2834 | 1.377 | 0.3827 | No | ||
| 35 | BACH2 | 3218 | 1.249 | 0.3702 | No | ||
| 36 | PREB | 3499 | 1.159 | 0.3627 | No | ||
| 37 | PCDHGC5 | 3603 | 1.123 | 0.3645 | No | ||
| 38 | MLX | 3931 | 1.021 | 0.3535 | No | ||
| 39 | DNMT3A | 4414 | 0.894 | 0.3334 | No | ||
| 40 | RHOQ | 4433 | 0.888 | 0.3382 | No | ||
| 41 | PRX | 4922 | 0.766 | 0.3169 | No | ||
| 42 | SMCR8 | 5065 | 0.732 | 0.3140 | No | ||
| 43 | AXL | 5194 | 0.704 | 0.3117 | No | ||
| 44 | GDAP2 | 5256 | 0.686 | 0.3129 | No | ||
| 45 | PCDHGA6 | 5259 | 0.686 | 0.3173 | No | ||
| 46 | PCDHGA10 | 5327 | 0.668 | 0.3181 | No | ||
| 47 | WDR1 | 5688 | 0.594 | 0.3025 | No | ||
| 48 | STC1 | 5791 | 0.573 | 0.3008 | No | ||
| 49 | PCDHGB7 | 5903 | 0.548 | 0.2984 | No | ||
| 50 | PCDHGA5 | 6068 | 0.510 | 0.2929 | No | ||
| 51 | ITGB3 | 6344 | 0.449 | 0.2810 | No | ||
| 52 | DLL4 | 6402 | 0.436 | 0.2807 | No | ||
| 53 | GOLGA7 | 6449 | 0.423 | 0.2810 | No | ||
| 54 | PCDHGC4 | 6512 | 0.410 | 0.2804 | No | ||
| 55 | CHCHD3 | 6811 | 0.346 | 0.2665 | No | ||
| 56 | PCDHGA4 | 7419 | 0.215 | 0.2352 | No | ||
| 57 | CD34 | 7811 | 0.146 | 0.2150 | No | ||
| 58 | PCDHGB2 | 8199 | 0.074 | 0.1946 | No | ||
| 59 | HNRPAB | 8309 | 0.054 | 0.1890 | No | ||
| 60 | GPRC5B | 8467 | 0.016 | 0.1806 | No | ||
| 61 | TWIST1 | 8516 | 0.006 | 0.1781 | No | ||
| 62 | MOBKL2B | 8518 | 0.006 | 0.1781 | No | ||
| 63 | PRR3 | 8603 | -0.012 | 0.1736 | No | ||
| 64 | ST3GAL4 | 8960 | -0.084 | 0.1549 | No | ||
| 65 | VAMP5 | 9009 | -0.093 | 0.1529 | No | ||
| 66 | ANK2 | 9106 | -0.113 | 0.1485 | No | ||
| 67 | RAB15 | 9161 | -0.126 | 0.1464 | No | ||
| 68 | EIF2C1 | 9211 | -0.138 | 0.1447 | No | ||
| 69 | PCDHGA3 | 9450 | -0.188 | 0.1331 | No | ||
| 70 | CSNK2A1 | 9542 | -0.208 | 0.1295 | No | ||
| 71 | ELAVL1 | 9576 | -0.215 | 0.1291 | No | ||
| 72 | LZTFL1 | 9729 | -0.249 | 0.1225 | No | ||
| 73 | PCDHGA9 | 9783 | -0.262 | 0.1214 | No | ||
| 74 | PCDHGB6 | 9966 | -0.299 | 0.1135 | No | ||
| 75 | HMGN1 | 9986 | -0.304 | 0.1145 | No | ||
| 76 | MEOX2 | 10027 | -0.314 | 0.1144 | No | ||
| 77 | PSME3 | 10728 | -0.459 | 0.0796 | No | ||
| 78 | PCDHGA8 | 10876 | -0.490 | 0.0749 | No | ||
| 79 | ETV5 | 11242 | -0.563 | 0.0588 | No | ||
| 80 | GDF8 | 11461 | -0.608 | 0.0511 | No | ||
| 81 | PCDHGB4 | 11513 | -0.617 | 0.0523 | No | ||
| 82 | PCDHGB5 | 12004 | -0.707 | 0.0305 | No | ||
| 83 | SPRY2 | 12080 | -0.720 | 0.0312 | No | ||
| 84 | CUTL2 | 12226 | -0.753 | 0.0283 | No | ||
| 85 | SALL2 | 12270 | -0.762 | 0.0310 | No | ||
| 86 | PCDHGA2 | 12342 | -0.780 | 0.0323 | No | ||
| 87 | SULF1 | 12850 | -0.889 | 0.0107 | No | ||
| 88 | HOXA1 | 13217 | -0.969 | -0.0027 | No | ||
| 89 | PCDHGA1 | 14272 | -1.212 | -0.0517 | No | ||
| 90 | RND1 | 14941 | -1.373 | -0.0788 | No | ||
| 91 | SYT7 | 14976 | -1.385 | -0.0715 | No | ||
| 92 | RHOT1 | 15040 | -1.397 | -0.0658 | No | ||
| 93 | SEPT4 | 15113 | -1.414 | -0.0604 | No | ||
| 94 | ARID5B | 15540 | -1.544 | -0.0733 | No | ||
| 95 | DDT | 15817 | -1.631 | -0.0775 | No | ||
| 96 | PCDHGA7 | 15914 | -1.668 | -0.0717 | No | ||
| 97 | GRK5 | 15943 | -1.676 | -0.0622 | No | ||
| 98 | SGCD | 16315 | -1.804 | -0.0704 | No | ||
| 99 | MAPK1 | 16496 | -1.873 | -0.0679 | No | ||
| 100 | MKLN1 | 16561 | -1.899 | -0.0588 | No | ||
| 101 | ING3 | 16650 | -1.940 | -0.0509 | No | ||
| 102 | SDK2 | 17022 | -2.118 | -0.0570 | No | ||
| 103 | TCF12 | 17170 | -2.211 | -0.0504 | No | ||
| 104 | RALA | 17413 | -2.375 | -0.0479 | No | ||
| 105 | ST8SIA2 | 17456 | -2.404 | -0.0344 | No | ||
| 106 | SLC30A4 | 17964 | -2.847 | -0.0431 | No | ||
| 107 | ZMYND11 | 18100 | -3.032 | -0.0305 | No | ||
| 108 | ACACA | 18291 | -3.340 | -0.0189 | No | ||
| 109 | NR4A2 | 18597 | -5.541 | 0.0010 | No |
