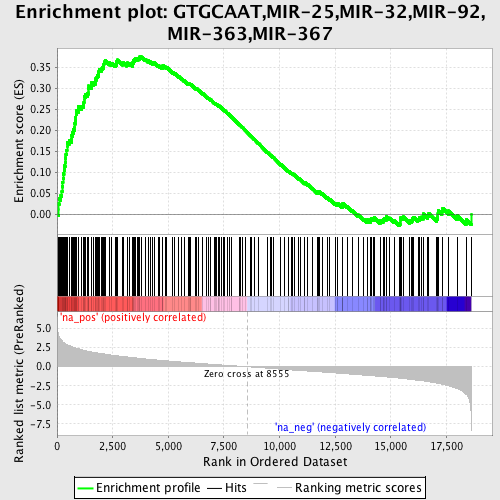
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | GTGCAAT,MIR-25,MIR-32,MIR-92,MIR-363,MIR-367 |
| Enrichment Score (ES) | 0.37436038 |
| Normalized Enrichment Score (NES) | 1.9258012 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.059437033 |
| FWER p-Value | 0.061 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FNBP4 | 57 | 4.178 | 0.0114 | Yes | ||
| 2 | BCL11A | 59 | 4.158 | 0.0258 | Yes | ||
| 3 | CIC | 90 | 3.897 | 0.0377 | Yes | ||
| 4 | RAB14 | 169 | 3.550 | 0.0457 | Yes | ||
| 5 | BRMS1L | 216 | 3.385 | 0.0550 | Yes | ||
| 6 | TRAK2 | 228 | 3.341 | 0.0660 | Yes | ||
| 7 | OGT | 242 | 3.281 | 0.0767 | Yes | ||
| 8 | RPS6KA4 | 265 | 3.219 | 0.0866 | Yes | ||
| 9 | SLC9A1 | 282 | 3.186 | 0.0968 | Yes | ||
| 10 | PCTK1 | 310 | 3.130 | 0.1062 | Yes | ||
| 11 | FBXW7 | 335 | 3.077 | 0.1156 | Yes | ||
| 12 | REV3L | 361 | 3.017 | 0.1247 | Yes | ||
| 13 | SLC38A2 | 374 | 2.993 | 0.1344 | Yes | ||
| 14 | KIF5B | 395 | 2.968 | 0.1436 | Yes | ||
| 15 | TRAF3 | 428 | 2.923 | 0.1520 | Yes | ||
| 16 | PIP5K3 | 446 | 2.900 | 0.1612 | Yes | ||
| 17 | NFAT5 | 451 | 2.890 | 0.1710 | Yes | ||
| 18 | PIP5K1C | 539 | 2.776 | 0.1759 | Yes | ||
| 19 | MYH9 | 644 | 2.638 | 0.1794 | Yes | ||
| 20 | MCL1 | 659 | 2.620 | 0.1877 | Yes | ||
| 21 | PPP1R12A | 694 | 2.591 | 0.1949 | Yes | ||
| 22 | ITPR1 | 728 | 2.554 | 0.2019 | Yes | ||
| 23 | PAX3 | 789 | 2.490 | 0.2073 | Yes | ||
| 24 | PER2 | 796 | 2.484 | 0.2156 | Yes | ||
| 25 | BTBD12 | 811 | 2.472 | 0.2234 | Yes | ||
| 26 | PTPRO | 838 | 2.449 | 0.2305 | Yes | ||
| 27 | MYO18A | 849 | 2.438 | 0.2384 | Yes | ||
| 28 | NSMAF | 862 | 2.428 | 0.2462 | Yes | ||
| 29 | TOB1 | 948 | 2.352 | 0.2498 | Yes | ||
| 30 | KLF2 | 966 | 2.341 | 0.2570 | Yes | ||
| 31 | HERC2 | 1108 | 2.230 | 0.2570 | Yes | ||
| 32 | BAZ2A | 1182 | 2.168 | 0.2606 | Yes | ||
| 33 | RSBN1 | 1202 | 2.156 | 0.2670 | Yes | ||
| 34 | FMN2 | 1212 | 2.147 | 0.2740 | Yes | ||
| 35 | PITPNC1 | 1216 | 2.142 | 0.2813 | Yes | ||
| 36 | VPS54 | 1277 | 2.100 | 0.2853 | Yes | ||
| 37 | ATP2A2 | 1370 | 2.038 | 0.2874 | Yes | ||
| 38 | CNTN4 | 1398 | 2.020 | 0.2929 | Yes | ||
| 39 | DOCK9 | 1400 | 2.020 | 0.2999 | Yes | ||
| 40 | NFIX | 1429 | 2.005 | 0.3053 | Yes | ||
| 41 | P2RY13 | 1537 | 1.947 | 0.3062 | Yes | ||
| 42 | ADAM19 | 1542 | 1.943 | 0.3128 | Yes | ||
| 43 | DSCAML1 | 1655 | 1.882 | 0.3132 | Yes | ||
| 44 | RNF44 | 1722 | 1.842 | 0.3160 | Yes | ||
| 45 | WBSCR18 | 1730 | 1.839 | 0.3220 | Yes | ||
| 46 | LYST | 1785 | 1.817 | 0.3254 | Yes | ||
| 47 | RGS17 | 1806 | 1.807 | 0.3306 | Yes | ||
| 48 | POLS | 1852 | 1.782 | 0.3343 | Yes | ||
| 49 | TACC2 | 1876 | 1.773 | 0.3392 | Yes | ||
| 50 | RNF38 | 1885 | 1.766 | 0.3449 | Yes | ||
| 51 | SSFA2 | 1980 | 1.719 | 0.3458 | Yes | ||
| 52 | CPEB3 | 2025 | 1.702 | 0.3493 | Yes | ||
| 53 | NELF | 2078 | 1.683 | 0.3523 | Yes | ||
| 54 | KLF4 | 2098 | 1.675 | 0.3571 | Yes | ||
| 55 | CSMD3 | 2108 | 1.668 | 0.3624 | Yes | ||
| 56 | POLK | 2161 | 1.645 | 0.3653 | Yes | ||
| 57 | CREB1 | 2352 | 1.562 | 0.3604 | Yes | ||
| 58 | SRPR | 2464 | 1.518 | 0.3596 | Yes | ||
| 59 | ROBO2 | 2635 | 1.450 | 0.3554 | Yes | ||
| 60 | SNF1LK | 2669 | 1.442 | 0.3586 | Yes | ||
| 61 | E2F3 | 2680 | 1.433 | 0.3630 | Yes | ||
| 62 | TSGA14 | 2699 | 1.429 | 0.3670 | Yes | ||
| 63 | CPEB2 | 2925 | 1.342 | 0.3595 | Yes | ||
| 64 | TOB2 | 2995 | 1.321 | 0.3603 | Yes | ||
| 65 | RBPMS2 | 3141 | 1.277 | 0.3568 | Yes | ||
| 66 | MAN2A1 | 3165 | 1.268 | 0.3600 | Yes | ||
| 67 | ATRX | 3275 | 1.228 | 0.3583 | Yes | ||
| 68 | IXL | 3394 | 1.194 | 0.3561 | Yes | ||
| 69 | ADM | 3398 | 1.192 | 0.3600 | Yes | ||
| 70 | DDX3Y | 3420 | 1.185 | 0.3630 | Yes | ||
| 71 | SERTAD2 | 3432 | 1.181 | 0.3665 | Yes | ||
| 72 | PHTF2 | 3489 | 1.161 | 0.3675 | Yes | ||
| 73 | PLEKHA6 | 3523 | 1.152 | 0.3697 | Yes | ||
| 74 | RFX1 | 3595 | 1.126 | 0.3698 | Yes | ||
| 75 | SGPP1 | 3650 | 1.109 | 0.3707 | Yes | ||
| 76 | CREB3L2 | 3696 | 1.092 | 0.3720 | Yes | ||
| 77 | HAND2 | 3723 | 1.083 | 0.3744 | Yes | ||
| 78 | TSC1 | 3795 | 1.062 | 0.3742 | No | ||
| 79 | KLHL18 | 3954 | 1.014 | 0.3691 | No | ||
| 80 | HCN2 | 4090 | 0.979 | 0.3652 | No | ||
| 81 | USP28 | 4200 | 0.948 | 0.3626 | No | ||
| 82 | OAZ3 | 4307 | 0.923 | 0.3600 | No | ||
| 83 | SOCS5 | 4360 | 0.910 | 0.3603 | No | ||
| 84 | CXCL5 | 4544 | 0.857 | 0.3534 | No | ||
| 85 | CD2AP | 4612 | 0.843 | 0.3527 | No | ||
| 86 | IQWD1 | 4730 | 0.813 | 0.3491 | No | ||
| 87 | PTGER4 | 4747 | 0.809 | 0.3511 | No | ||
| 88 | TCF21 | 4756 | 0.806 | 0.3534 | No | ||
| 89 | APRIN | 4861 | 0.780 | 0.3505 | No | ||
| 90 | PPP1R12C | 4918 | 0.766 | 0.3501 | No | ||
| 91 | GATA6 | 5190 | 0.705 | 0.3378 | No | ||
| 92 | CNNM4 | 5281 | 0.681 | 0.3353 | No | ||
| 93 | HIVEP1 | 5472 | 0.636 | 0.3272 | No | ||
| 94 | SLC32A1 | 5591 | 0.612 | 0.3229 | No | ||
| 95 | EGR2 | 5710 | 0.590 | 0.3185 | No | ||
| 96 | FMR1 | 5913 | 0.545 | 0.3094 | No | ||
| 97 | DUSP10 | 5933 | 0.541 | 0.3103 | No | ||
| 98 | DAB2IP | 5981 | 0.528 | 0.3096 | No | ||
| 99 | NRF1 | 6010 | 0.521 | 0.3099 | No | ||
| 100 | BCAT2 | 6233 | 0.474 | 0.2994 | No | ||
| 101 | HOXB8 | 6257 | 0.469 | 0.2998 | No | ||
| 102 | TEAD1 | 6364 | 0.442 | 0.2956 | No | ||
| 103 | SMAD7 | 6544 | 0.403 | 0.2873 | No | ||
| 104 | NLK | 6725 | 0.363 | 0.2788 | No | ||
| 105 | ARMC1 | 6792 | 0.349 | 0.2764 | No | ||
| 106 | DDX3X | 6889 | 0.328 | 0.2723 | No | ||
| 107 | ARRDC4 | 6916 | 0.322 | 0.2720 | No | ||
| 108 | GATA2 | 7089 | 0.283 | 0.2637 | No | ||
| 109 | RHPN2 | 7100 | 0.281 | 0.2641 | No | ||
| 110 | GOLGA4 | 7150 | 0.272 | 0.2624 | No | ||
| 111 | GDF11 | 7264 | 0.249 | 0.2571 | No | ||
| 112 | ITGAV | 7269 | 0.248 | 0.2577 | No | ||
| 113 | ITGA5 | 7284 | 0.245 | 0.2578 | No | ||
| 114 | PCDH11X | 7371 | 0.226 | 0.2539 | No | ||
| 115 | HAS3 | 7477 | 0.204 | 0.2489 | No | ||
| 116 | FZD10 | 7539 | 0.193 | 0.2463 | No | ||
| 117 | SDC2 | 7649 | 0.175 | 0.2410 | No | ||
| 118 | MEF2D | 7737 | 0.159 | 0.2368 | No | ||
| 119 | PIK3R3 | 7845 | 0.138 | 0.2315 | No | ||
| 120 | EDG1 | 7848 | 0.137 | 0.2318 | No | ||
| 121 | DNAJB9 | 8181 | 0.077 | 0.2141 | No | ||
| 122 | PIK3AP1 | 8190 | 0.075 | 0.2139 | No | ||
| 123 | PITPNA | 8244 | 0.066 | 0.2113 | No | ||
| 124 | COL12A1 | 8315 | 0.052 | 0.2076 | No | ||
| 125 | SIM2 | 8460 | 0.017 | 0.1999 | No | ||
| 126 | EPHA8 | 8670 | -0.026 | 0.1886 | No | ||
| 127 | SESN3 | 8749 | -0.040 | 0.1845 | No | ||
| 128 | FASLG | 8851 | -0.060 | 0.1792 | No | ||
| 129 | WRNIP1 | 9064 | -0.104 | 0.1681 | No | ||
| 130 | MYO1B | 9466 | -0.191 | 0.1470 | No | ||
| 131 | FHL2 | 9584 | -0.217 | 0.1414 | No | ||
| 132 | PRKCE | 9636 | -0.228 | 0.1394 | No | ||
| 133 | ANP32E | 9705 | -0.242 | 0.1365 | No | ||
| 134 | HHIP | 10062 | -0.324 | 0.1183 | No | ||
| 135 | FBN1 | 10232 | -0.359 | 0.1104 | No | ||
| 136 | BSN | 10391 | -0.393 | 0.1032 | No | ||
| 137 | DSC2 | 10422 | -0.399 | 0.1029 | No | ||
| 138 | LHFPL2 | 10531 | -0.418 | 0.0985 | No | ||
| 139 | SYN2 | 10601 | -0.434 | 0.0963 | No | ||
| 140 | ADAM10 | 10666 | -0.446 | 0.0943 | No | ||
| 141 | RAD21 | 10861 | -0.486 | 0.0855 | No | ||
| 142 | SOX11 | 10920 | -0.499 | 0.0841 | No | ||
| 143 | TRAM2 | 11135 | -0.541 | 0.0743 | No | ||
| 144 | IBTK | 11137 | -0.542 | 0.0761 | No | ||
| 145 | SOX4 | 11266 | -0.568 | 0.0712 | No | ||
| 146 | COL1A2 | 11466 | -0.608 | 0.0625 | No | ||
| 147 | SMAD6 | 11706 | -0.651 | 0.0517 | No | ||
| 148 | CD69 | 11724 | -0.656 | 0.0531 | No | ||
| 149 | BTG2 | 11744 | -0.658 | 0.0543 | No | ||
| 150 | PAFAH1B1 | 11781 | -0.667 | 0.0547 | No | ||
| 151 | ATXN3 | 11927 | -0.695 | 0.0492 | No | ||
| 152 | CBLN4 | 12143 | -0.733 | 0.0401 | No | ||
| 153 | XYLT2 | 12263 | -0.761 | 0.0363 | No | ||
| 154 | SLC12A5 | 12492 | -0.809 | 0.0267 | No | ||
| 155 | STK39 | 12603 | -0.837 | 0.0236 | No | ||
| 156 | TULP4 | 12614 | -0.840 | 0.0260 | No | ||
| 157 | NFIA | 12805 | -0.880 | 0.0187 | No | ||
| 158 | NOX4 | 12834 | -0.886 | 0.0203 | No | ||
| 159 | ARHGEF17 | 12835 | -0.887 | 0.0234 | No | ||
| 160 | BCL2L11 | 12844 | -0.888 | 0.0260 | No | ||
| 161 | DNAJB12 | 13031 | -0.928 | 0.0191 | No | ||
| 162 | RNF4 | 13260 | -0.979 | 0.0101 | No | ||
| 163 | PRDM13 | 13534 | -1.037 | -0.0011 | No | ||
| 164 | DLGAP2 | 13786 | -1.095 | -0.0109 | No | ||
| 165 | DPP10 | 13945 | -1.136 | -0.0156 | No | ||
| 166 | TEF | 13964 | -1.141 | -0.0126 | No | ||
| 167 | UGP2 | 14074 | -1.163 | -0.0145 | No | ||
| 168 | GRIA3 | 14099 | -1.169 | -0.0117 | No | ||
| 169 | CXXC5 | 14121 | -1.176 | -0.0088 | No | ||
| 170 | MAP2K4 | 14231 | -1.203 | -0.0105 | No | ||
| 171 | MYLIP | 14243 | -1.207 | -0.0069 | No | ||
| 172 | SLC24A3 | 14519 | -1.274 | -0.0175 | No | ||
| 173 | GFPT2 | 14530 | -1.276 | -0.0136 | No | ||
| 174 | INSIG1 | 14651 | -1.302 | -0.0156 | No | ||
| 175 | RAP1B | 14660 | -1.304 | -0.0115 | No | ||
| 176 | EXOC5 | 14716 | -1.317 | -0.0099 | No | ||
| 177 | SLC17A6 | 14788 | -1.335 | -0.0091 | No | ||
| 178 | HAND1 | 14790 | -1.336 | -0.0045 | No | ||
| 179 | BMPR2 | 14947 | -1.375 | -0.0082 | No | ||
| 180 | RAB23 | 15143 | -1.423 | -0.0139 | No | ||
| 181 | CEBPA | 15389 | -1.498 | -0.0220 | No | ||
| 182 | NFIB | 15419 | -1.508 | -0.0184 | No | ||
| 183 | NR4A3 | 15420 | -1.508 | -0.0131 | No | ||
| 184 | LUZP1 | 15424 | -1.508 | -0.0080 | No | ||
| 185 | EOMES | 15486 | -1.529 | -0.0061 | No | ||
| 186 | CCNE2 | 15577 | -1.555 | -0.0056 | No | ||
| 187 | MOAP1 | 15856 | -1.648 | -0.0149 | No | ||
| 188 | GAP43 | 15929 | -1.671 | -0.0130 | No | ||
| 189 | PCAF | 15978 | -1.689 | -0.0098 | No | ||
| 190 | TCF2 | 16026 | -1.706 | -0.0064 | No | ||
| 191 | RNF141 | 16221 | -1.772 | -0.0108 | No | ||
| 192 | ZDHHC5 | 16275 | -1.790 | -0.0075 | No | ||
| 193 | CCNC | 16363 | -1.821 | -0.0059 | No | ||
| 194 | MARK1 | 16453 | -1.851 | -0.0043 | No | ||
| 195 | EDEM1 | 16467 | -1.859 | 0.0014 | No | ||
| 196 | PGAM1 | 16660 | -1.946 | -0.0022 | No | ||
| 197 | RGS3 | 16710 | -1.967 | 0.0019 | No | ||
| 198 | LATS2 | 17066 | -2.144 | -0.0099 | No | ||
| 199 | SCN3A | 17112 | -2.174 | -0.0048 | No | ||
| 200 | DYNLT3 | 17118 | -2.177 | 0.0025 | No | ||
| 201 | LBX1 | 17138 | -2.183 | 0.0090 | No | ||
| 202 | ARRDC3 | 17301 | -2.290 | 0.0082 | No | ||
| 203 | SERTAD3 | 17337 | -2.317 | 0.0143 | No | ||
| 204 | NPTX1 | 17603 | -2.511 | 0.0086 | No | ||
| 205 | ADCY3 | 17986 | -2.884 | -0.0021 | No | ||
| 206 | CDKN1C | 18384 | -3.606 | -0.0112 | No | ||
| 207 | ELOVL4 | 18613 | -6.841 | 0.0002 | No |
