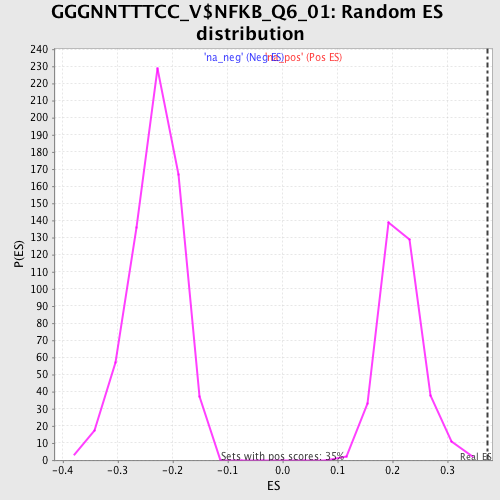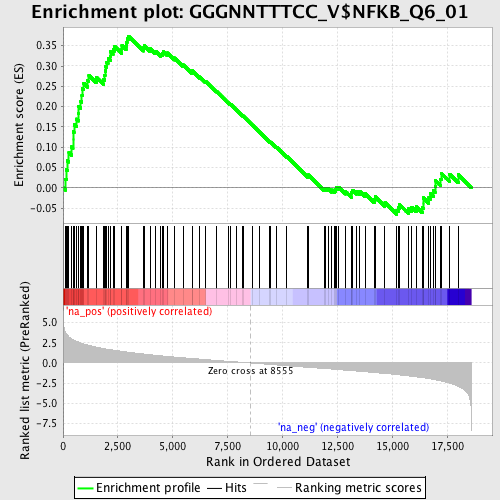
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | GGGNNTTTCC_V$NFKB_Q6_01 |
| Enrichment Score (ES) | 0.37360814 |
| Normalized Enrichment Score (NES) | 1.7355875 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07721258 |
| FWER p-Value | 0.326 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BNC2 | 107 | 3.801 | 0.0213 | Yes | ||
| 2 | HSPG2 | 146 | 3.620 | 0.0450 | Yes | ||
| 3 | CNTF | 195 | 3.445 | 0.0670 | Yes | ||
| 4 | RPS6KA4 | 265 | 3.219 | 0.0862 | Yes | ||
| 5 | ARID1A | 388 | 2.976 | 0.1008 | Yes | ||
| 6 | NFAT5 | 451 | 2.890 | 0.1181 | Yes | ||
| 7 | RASSF2 | 452 | 2.885 | 0.1386 | Yes | ||
| 8 | TP53 | 501 | 2.825 | 0.1561 | Yes | ||
| 9 | TCEA2 | 610 | 2.691 | 0.1695 | Yes | ||
| 10 | NFKB2 | 704 | 2.580 | 0.1828 | Yes | ||
| 11 | PPP2R5E | 714 | 2.568 | 0.2007 | Yes | ||
| 12 | TPM3 | 805 | 2.478 | 0.2135 | Yes | ||
| 13 | BMP2K | 856 | 2.431 | 0.2281 | Yes | ||
| 14 | SLC25A18 | 871 | 2.422 | 0.2446 | Yes | ||
| 15 | CD86 | 940 | 2.360 | 0.2577 | Yes | ||
| 16 | TNIP1 | 1121 | 2.216 | 0.2638 | Yes | ||
| 17 | UBE2H | 1175 | 2.174 | 0.2764 | Yes | ||
| 18 | TNFSF15 | 1513 | 1.960 | 0.2722 | Yes | ||
| 19 | LTB | 1849 | 1.784 | 0.2668 | Yes | ||
| 20 | SMPD3 | 1886 | 1.766 | 0.2774 | Yes | ||
| 21 | UPF2 | 1913 | 1.752 | 0.2885 | Yes | ||
| 22 | SH3KBP1 | 1939 | 1.739 | 0.2996 | Yes | ||
| 23 | IER5 | 1988 | 1.716 | 0.3092 | Yes | ||
| 24 | MYADM | 2048 | 1.693 | 0.3181 | Yes | ||
| 25 | LASP1 | 2155 | 1.648 | 0.3241 | Yes | ||
| 26 | BLR1 | 2169 | 1.641 | 0.3351 | Yes | ||
| 27 | RBMS1 | 2276 | 1.594 | 0.3407 | Yes | ||
| 28 | CREB1 | 2352 | 1.562 | 0.3478 | Yes | ||
| 29 | STAT6 | 2672 | 1.439 | 0.3408 | Yes | ||
| 30 | E2F3 | 2680 | 1.433 | 0.3507 | Yes | ||
| 31 | IL6 | 2872 | 1.363 | 0.3501 | Yes | ||
| 32 | CCND2 | 2900 | 1.351 | 0.3582 | Yes | ||
| 33 | JAK3 | 2916 | 1.346 | 0.3670 | Yes | ||
| 34 | TOM1L1 | 2970 | 1.330 | 0.3736 | Yes | ||
| 35 | RYBP | 3651 | 1.109 | 0.3448 | No | ||
| 36 | NFKBIA | 3692 | 1.094 | 0.3504 | No | ||
| 37 | BIRC3 | 3973 | 1.009 | 0.3425 | No | ||
| 38 | TNKS1BP1 | 4208 | 0.946 | 0.3366 | No | ||
| 39 | SP6 | 4458 | 0.881 | 0.3294 | No | ||
| 40 | BATF | 4537 | 0.860 | 0.3313 | No | ||
| 41 | SLAMF8 | 4562 | 0.854 | 0.3361 | No | ||
| 42 | UBD | 4742 | 0.811 | 0.3322 | No | ||
| 43 | NRIP2 | 5068 | 0.731 | 0.3199 | No | ||
| 44 | HIVEP1 | 5472 | 0.636 | 0.3027 | No | ||
| 45 | DCAMKL1 | 5880 | 0.553 | 0.2846 | No | ||
| 46 | TRIM47 | 5888 | 0.552 | 0.2882 | No | ||
| 47 | CCL5 | 6229 | 0.475 | 0.2732 | No | ||
| 48 | EHD1 | 6491 | 0.414 | 0.2621 | No | ||
| 49 | CYLD | 6978 | 0.308 | 0.2380 | No | ||
| 50 | KCNS3 | 7541 | 0.193 | 0.2090 | No | ||
| 51 | REL | 7630 | 0.179 | 0.2056 | No | ||
| 52 | TFEC | 7883 | 0.130 | 0.1929 | No | ||
| 53 | PFN1 | 8160 | 0.080 | 0.1786 | No | ||
| 54 | LPO | 8200 | 0.074 | 0.1770 | No | ||
| 55 | FUT7 | 8636 | -0.020 | 0.1536 | No | ||
| 56 | MYST2 | 8952 | -0.081 | 0.1372 | No | ||
| 57 | BCL6B | 9428 | -0.183 | 0.1128 | No | ||
| 58 | LAMA1 | 9473 | -0.193 | 0.1118 | No | ||
| 59 | BCL3 | 9728 | -0.249 | 0.0999 | No | ||
| 60 | IL1RN | 10194 | -0.349 | 0.0773 | No | ||
| 61 | SLC6A12 | 11155 | -0.545 | 0.0293 | No | ||
| 62 | MSX1 | 11182 | -0.551 | 0.0318 | No | ||
| 63 | PTGES | 11897 | -0.689 | -0.0018 | No | ||
| 64 | TNFRSF1B | 11967 | -0.701 | -0.0005 | No | ||
| 65 | MADCAM1 | 12073 | -0.719 | -0.0011 | No | ||
| 66 | UBE2M | 12253 | -0.760 | -0.0053 | No | ||
| 67 | FKHL18 | 12379 | -0.787 | -0.0065 | No | ||
| 68 | EDG5 | 12421 | -0.795 | -0.0030 | No | ||
| 69 | GATA4 | 12454 | -0.801 | 0.0009 | No | ||
| 70 | POU2F3 | 12561 | -0.827 | 0.0011 | No | ||
| 71 | ETV1 | 12869 | -0.893 | -0.0091 | No | ||
| 72 | HOXA4 | 13148 | -0.952 | -0.0173 | No | ||
| 73 | GREM1 | 13151 | -0.952 | -0.0107 | No | ||
| 74 | ALG6 | 13187 | -0.961 | -0.0057 | No | ||
| 75 | FHL3 | 13371 | -1.003 | -0.0084 | No | ||
| 76 | FGF1 | 13522 | -1.034 | -0.0092 | No | ||
| 77 | RELB | 13763 | -1.089 | -0.0144 | No | ||
| 78 | TNFRSF9 | 14191 | -1.194 | -0.0289 | No | ||
| 79 | CNTNAP1 | 14217 | -1.199 | -0.0217 | No | ||
| 80 | MT2A | 14669 | -1.306 | -0.0368 | No | ||
| 81 | FTHL17 | 15207 | -1.439 | -0.0555 | No | ||
| 82 | TLX1 | 15280 | -1.459 | -0.0490 | No | ||
| 83 | ADMR | 15324 | -1.474 | -0.0408 | No | ||
| 84 | CSF1R | 15741 | -1.608 | -0.0518 | No | ||
| 85 | NR2F2 | 15883 | -1.657 | -0.0476 | No | ||
| 86 | SLC12A2 | 16093 | -1.732 | -0.0466 | No | ||
| 87 | MSC | 16369 | -1.823 | -0.0484 | No | ||
| 88 | RAP2C | 16413 | -1.837 | -0.0377 | No | ||
| 89 | SOX5 | 16414 | -1.837 | -0.0246 | No | ||
| 90 | WNT10A | 16669 | -1.949 | -0.0244 | No | ||
| 91 | PCSK2 | 16737 | -1.978 | -0.0139 | No | ||
| 92 | GEM | 16878 | -2.048 | -0.0069 | No | ||
| 93 | ENO3 | 16955 | -2.084 | 0.0038 | No | ||
| 94 | NTRK3 | 16980 | -2.096 | 0.0175 | No | ||
| 95 | IPO4 | 17200 | -2.228 | 0.0215 | No | ||
| 96 | PLXNB1 | 17237 | -2.250 | 0.0356 | No | ||
| 97 | IL2 | 17618 | -2.529 | 0.0331 | No | ||
| 98 | C1QL1 | 18013 | -2.910 | 0.0326 | No |