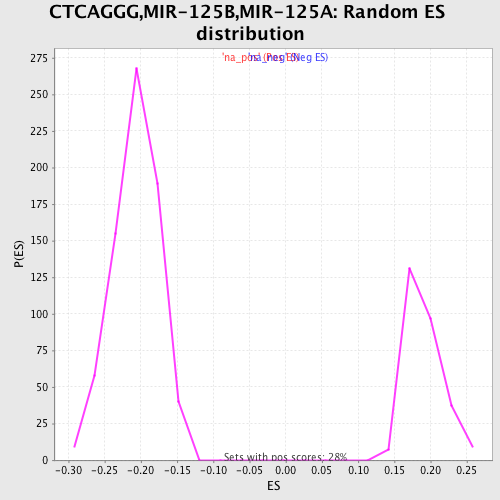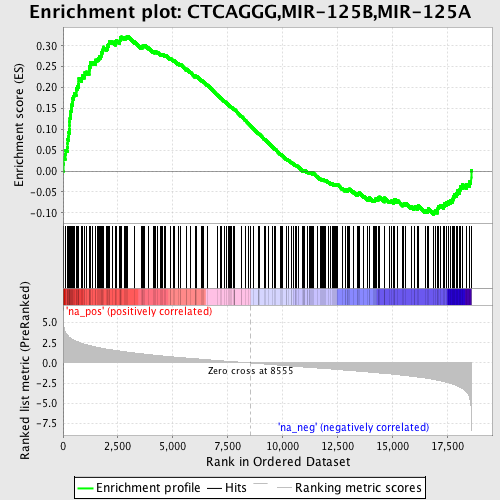
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CTCAGGG,MIR-125B,MIR-125A |
| Enrichment Score (ES) | 0.32310003 |
| Normalized Enrichment Score (NES) | 1.6952697 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.06775216 |
| FWER p-Value | 0.432 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GALNT14 | 5 | 5.866 | 0.0171 | Yes | ||
| 2 | TRAPPC6B | 13 | 5.247 | 0.0323 | Yes | ||
| 3 | SIRT7 | 91 | 3.897 | 0.0396 | Yes | ||
| 4 | MAP3K11 | 104 | 3.838 | 0.0503 | Yes | ||
| 5 | OLFML2A | 189 | 3.481 | 0.0561 | Yes | ||
| 6 | PAPOLA | 205 | 3.413 | 0.0654 | Yes | ||
| 7 | ARID3B | 210 | 3.393 | 0.0752 | Yes | ||
| 8 | BRPF1 | 247 | 3.273 | 0.0829 | Yes | ||
| 9 | TGOLN2 | 259 | 3.233 | 0.0919 | Yes | ||
| 10 | NIN | 285 | 3.177 | 0.1000 | Yes | ||
| 11 | NRM | 299 | 3.147 | 0.1086 | Yes | ||
| 12 | ZBTB7A | 304 | 3.137 | 0.1177 | Yes | ||
| 13 | PCTK1 | 310 | 3.130 | 0.1267 | Yes | ||
| 14 | CBX7 | 334 | 3.077 | 0.1345 | Yes | ||
| 15 | PLAGL2 | 353 | 3.031 | 0.1425 | Yes | ||
| 16 | GGA2 | 368 | 3.006 | 0.1507 | Yes | ||
| 17 | NUP210 | 389 | 2.975 | 0.1584 | Yes | ||
| 18 | SYVN1 | 427 | 2.923 | 0.1650 | Yes | ||
| 19 | MAPK14 | 438 | 2.913 | 0.1731 | Yes | ||
| 20 | STAT3 | 479 | 2.857 | 0.1794 | Yes | ||
| 21 | SUV420H2 | 498 | 2.830 | 0.1868 | Yes | ||
| 22 | RNF40 | 592 | 2.704 | 0.1898 | Yes | ||
| 23 | FAM62A | 594 | 2.702 | 0.1977 | Yes | ||
| 24 | MCL1 | 659 | 2.620 | 0.2020 | Yes | ||
| 25 | RPS6KA1 | 701 | 2.582 | 0.2074 | Yes | ||
| 26 | GTPBP2 | 719 | 2.562 | 0.2141 | Yes | ||
| 27 | SRF | 721 | 2.560 | 0.2216 | Yes | ||
| 28 | MYO18A | 849 | 2.438 | 0.2219 | Yes | ||
| 29 | IL16 | 868 | 2.424 | 0.2281 | Yes | ||
| 30 | EIF2C2 | 969 | 2.339 | 0.2296 | Yes | ||
| 31 | USP52 | 994 | 2.324 | 0.2352 | Yes | ||
| 32 | SLC7A1 | 1080 | 2.262 | 0.2373 | Yes | ||
| 33 | BAZ2A | 1182 | 2.168 | 0.2382 | Yes | ||
| 34 | MKNK2 | 1184 | 2.166 | 0.2446 | Yes | ||
| 35 | EAF1 | 1194 | 2.159 | 0.2505 | Yes | ||
| 36 | KLF13 | 1232 | 2.131 | 0.2548 | Yes | ||
| 37 | NCOR2 | 1246 | 2.123 | 0.2604 | Yes | ||
| 38 | SLC39A9 | 1350 | 2.048 | 0.2608 | Yes | ||
| 39 | CASC3 | 1466 | 1.989 | 0.2604 | Yes | ||
| 40 | PODXL | 1474 | 1.987 | 0.2660 | Yes | ||
| 41 | BCL9L | 1547 | 1.941 | 0.2678 | Yes | ||
| 42 | PCSK7 | 1605 | 1.908 | 0.2703 | Yes | ||
| 43 | UBTD1 | 1645 | 1.886 | 0.2738 | Yes | ||
| 44 | RNF44 | 1722 | 1.842 | 0.2751 | Yes | ||
| 45 | APOBEC1 | 1733 | 1.838 | 0.2800 | Yes | ||
| 46 | PARP14 | 1755 | 1.826 | 0.2843 | Yes | ||
| 47 | TXNRD1 | 1789 | 1.815 | 0.2879 | Yes | ||
| 48 | HIC2 | 1816 | 1.801 | 0.2918 | Yes | ||
| 49 | SMURF1 | 1818 | 1.799 | 0.2971 | Yes | ||
| 50 | CORO2A | 1981 | 1.718 | 0.2933 | Yes | ||
| 51 | LNPEP | 2013 | 1.707 | 0.2967 | Yes | ||
| 52 | CPEB3 | 2025 | 1.702 | 0.3012 | Yes | ||
| 53 | ETS1 | 2076 | 1.684 | 0.3034 | Yes | ||
| 54 | CDC42SE1 | 2091 | 1.677 | 0.3076 | Yes | ||
| 55 | ESRRA | 2115 | 1.665 | 0.3113 | Yes | ||
| 56 | TLK2 | 2242 | 1.609 | 0.3092 | Yes | ||
| 57 | ABR | 2384 | 1.546 | 0.3061 | Yes | ||
| 58 | RBM7 | 2386 | 1.545 | 0.3107 | Yes | ||
| 59 | DCTN1 | 2441 | 1.525 | 0.3122 | Yes | ||
| 60 | ZNF76 | 2585 | 1.471 | 0.3088 | Yes | ||
| 61 | PCQAP | 2592 | 1.469 | 0.3128 | Yes | ||
| 62 | RHOT2 | 2603 | 1.465 | 0.3166 | Yes | ||
| 63 | HMGB3 | 2616 | 1.460 | 0.3203 | Yes | ||
| 64 | E2F3 | 2680 | 1.433 | 0.3211 | Yes | ||
| 65 | USP12 | 2793 | 1.393 | 0.3192 | Yes | ||
| 66 | PCGF6 | 2856 | 1.368 | 0.3198 | Yes | ||
| 67 | CNNM1 | 2909 | 1.348 | 0.3210 | Yes | ||
| 68 | SEMA4D | 2944 | 1.336 | 0.3231 | Yes | ||
| 69 | FBXW4 | 3262 | 1.232 | 0.3095 | No | ||
| 70 | ZFYVE1 | 3555 | 1.143 | 0.2970 | No | ||
| 71 | SLC4A10 | 3602 | 1.125 | 0.2978 | No | ||
| 72 | GOPC | 3633 | 1.115 | 0.2995 | No | ||
| 73 | SH3BP4 | 3681 | 1.097 | 0.3002 | No | ||
| 74 | VPS4B | 3720 | 1.084 | 0.3013 | No | ||
| 75 | SSTR3 | 3892 | 1.029 | 0.2951 | No | ||
| 76 | RTN2 | 4116 | 0.972 | 0.2858 | No | ||
| 77 | LIFR | 4178 | 0.954 | 0.2853 | No | ||
| 78 | DICER1 | 4210 | 0.946 | 0.2864 | No | ||
| 79 | SLC4A4 | 4290 | 0.929 | 0.2849 | No | ||
| 80 | RHOQ | 4433 | 0.888 | 0.2798 | No | ||
| 81 | ACCN2 | 4475 | 0.877 | 0.2802 | No | ||
| 82 | ASB13 | 4531 | 0.862 | 0.2797 | No | ||
| 83 | DAG1 | 4641 | 0.838 | 0.2763 | No | ||
| 84 | VANGL2 | 4682 | 0.827 | 0.2766 | No | ||
| 85 | RET | 4873 | 0.777 | 0.2685 | No | ||
| 86 | SMARCD2 | 4877 | 0.775 | 0.2706 | No | ||
| 87 | ELOVL6 | 5042 | 0.739 | 0.2639 | No | ||
| 88 | PELI2 | 5079 | 0.728 | 0.2641 | No | ||
| 89 | DVL3 | 5247 | 0.690 | 0.2571 | No | ||
| 90 | ABHD3 | 5343 | 0.664 | 0.2539 | No | ||
| 91 | SEMA4B | 5347 | 0.663 | 0.2557 | No | ||
| 92 | LRP4 | 5613 | 0.608 | 0.2430 | No | ||
| 93 | LRFN2 | 5621 | 0.607 | 0.2445 | No | ||
| 94 | STC1 | 5791 | 0.573 | 0.2370 | No | ||
| 95 | ATP10D | 6013 | 0.520 | 0.2265 | No | ||
| 96 | SLC35A4 | 6028 | 0.518 | 0.2273 | No | ||
| 97 | LBH | 6044 | 0.516 | 0.2280 | No | ||
| 98 | RAB8B | 6095 | 0.503 | 0.2267 | No | ||
| 99 | BAK1 | 6289 | 0.462 | 0.2176 | No | ||
| 100 | ITGB3 | 6344 | 0.449 | 0.2160 | No | ||
| 101 | DLL4 | 6402 | 0.436 | 0.2142 | No | ||
| 102 | ADAM9 | 6572 | 0.398 | 0.2062 | No | ||
| 103 | UBN1 | 7027 | 0.297 | 0.1823 | No | ||
| 104 | GRB10 | 7191 | 0.263 | 0.1743 | No | ||
| 105 | KLC2 | 7214 | 0.259 | 0.1738 | No | ||
| 106 | PTPN18 | 7351 | 0.232 | 0.1671 | No | ||
| 107 | TBX4 | 7427 | 0.214 | 0.1637 | No | ||
| 108 | KCNS3 | 7541 | 0.193 | 0.1581 | No | ||
| 109 | HOXB3 | 7595 | 0.185 | 0.1558 | No | ||
| 110 | APBA2BP | 7620 | 0.181 | 0.1550 | No | ||
| 111 | DPF2 | 7667 | 0.173 | 0.1530 | No | ||
| 112 | PCTP | 7777 | 0.152 | 0.1475 | No | ||
| 113 | LYPLA2 | 7792 | 0.149 | 0.1472 | No | ||
| 114 | CD34 | 7811 | 0.146 | 0.1466 | No | ||
| 115 | MYLK | 7818 | 0.144 | 0.1467 | No | ||
| 116 | PHOX2B | 7825 | 0.142 | 0.1468 | No | ||
| 117 | IER2 | 8125 | 0.086 | 0.1308 | No | ||
| 118 | FUT4 | 8130 | 0.085 | 0.1309 | No | ||
| 119 | ADD2 | 8299 | 0.056 | 0.1219 | No | ||
| 120 | BMF | 8450 | 0.020 | 0.1138 | No | ||
| 121 | SATB2 | 8555 | -0.000 | 0.1081 | No | ||
| 122 | UBE2R2 | 8689 | -0.030 | 0.1010 | No | ||
| 123 | IHPK1 | 8903 | -0.071 | 0.0896 | No | ||
| 124 | MYT1 | 8908 | -0.072 | 0.0896 | No | ||
| 125 | LFNG | 8924 | -0.075 | 0.0890 | No | ||
| 126 | MLF2 | 8941 | -0.079 | 0.0884 | No | ||
| 127 | SGPL1 | 9178 | -0.129 | 0.0759 | No | ||
| 128 | BAP1 | 9209 | -0.138 | 0.0747 | No | ||
| 129 | MMP11 | 9349 | -0.168 | 0.0676 | No | ||
| 130 | CSNK2A1 | 9542 | -0.208 | 0.0578 | No | ||
| 131 | TBC1D1 | 9623 | -0.225 | 0.0541 | No | ||
| 132 | ATXN1 | 9674 | -0.236 | 0.0521 | No | ||
| 133 | CACNB1 | 9922 | -0.289 | 0.0395 | No | ||
| 134 | ANPEP | 9960 | -0.298 | 0.0384 | No | ||
| 135 | BAI1 | 9982 | -0.303 | 0.0381 | No | ||
| 136 | RAB3D | 10172 | -0.343 | 0.0288 | No | ||
| 137 | SLC17A7 | 10201 | -0.351 | 0.0284 | No | ||
| 138 | NEU1 | 10278 | -0.370 | 0.0253 | No | ||
| 139 | BSN | 10391 | -0.393 | 0.0204 | No | ||
| 140 | SEMA4C | 10410 | -0.396 | 0.0206 | No | ||
| 141 | OSBPL9 | 10493 | -0.411 | 0.0173 | No | ||
| 142 | EIF5A2 | 10580 | -0.429 | 0.0139 | No | ||
| 143 | SYN2 | 10601 | -0.434 | 0.0141 | No | ||
| 144 | SLC8A2 | 10643 | -0.441 | 0.0132 | No | ||
| 145 | LACTB | 10729 | -0.459 | 0.0099 | No | ||
| 146 | SOX11 | 10920 | -0.499 | 0.0011 | No | ||
| 147 | UBE2L3 | 10935 | -0.503 | 0.0018 | No | ||
| 148 | SLITRK6 | 10951 | -0.505 | 0.0025 | No | ||
| 149 | ABHD6 | 10991 | -0.517 | 0.0019 | No | ||
| 150 | ST6GAL1 | 11016 | -0.522 | 0.0021 | No | ||
| 151 | CCNJ | 11129 | -0.540 | -0.0024 | No | ||
| 152 | NT5DC1 | 11157 | -0.545 | -0.0022 | No | ||
| 153 | ST8SIA4 | 11231 | -0.561 | -0.0045 | No | ||
| 154 | SORT1 | 11251 | -0.564 | -0.0039 | No | ||
| 155 | GPC4 | 11252 | -0.565 | -0.0022 | No | ||
| 156 | IBRDC2 | 11323 | -0.579 | -0.0043 | No | ||
| 157 | MXD4 | 11387 | -0.591 | -0.0060 | No | ||
| 158 | FCHSD1 | 11392 | -0.592 | -0.0044 | No | ||
| 159 | USP37 | 11579 | -0.628 | -0.0127 | No | ||
| 160 | GRIN2A | 11728 | -0.656 | -0.0188 | No | ||
| 161 | PAFAH1B1 | 11781 | -0.667 | -0.0197 | No | ||
| 162 | DDX42 | 11812 | -0.673 | -0.0193 | No | ||
| 163 | MAP2K7 | 11878 | -0.685 | -0.0208 | No | ||
| 164 | ETV6 | 11933 | -0.696 | -0.0217 | No | ||
| 165 | TOR2A | 11969 | -0.702 | -0.0215 | No | ||
| 166 | CGN | 12096 | -0.723 | -0.0262 | No | ||
| 167 | PHYHIP | 12202 | -0.749 | -0.0297 | No | ||
| 168 | KCNH7 | 12258 | -0.760 | -0.0305 | No | ||
| 169 | ATP5G2 | 12328 | -0.777 | -0.0319 | No | ||
| 170 | FKHL18 | 12379 | -0.787 | -0.0323 | No | ||
| 171 | CASP2 | 12400 | -0.791 | -0.0310 | No | ||
| 172 | SEMA4F | 12473 | -0.804 | -0.0326 | No | ||
| 173 | PPAT | 12489 | -0.809 | -0.0310 | No | ||
| 174 | SCARB1 | 12740 | -0.868 | -0.0420 | No | ||
| 175 | EMID1 | 12848 | -0.889 | -0.0452 | No | ||
| 176 | ZNF385 | 12860 | -0.891 | -0.0432 | No | ||
| 177 | PPP2R5C | 12959 | -0.911 | -0.0458 | No | ||
| 178 | PHC2 | 12969 | -0.914 | -0.0436 | No | ||
| 179 | PRDM1 | 13010 | -0.924 | -0.0430 | No | ||
| 180 | MAP3K10 | 13057 | -0.932 | -0.0428 | No | ||
| 181 | MIB1 | 13239 | -0.973 | -0.0497 | No | ||
| 182 | ENTPD4 | 13418 | -1.012 | -0.0564 | No | ||
| 183 | ABTB1 | 13430 | -1.015 | -0.0540 | No | ||
| 184 | ABCC5 | 13477 | -1.026 | -0.0535 | No | ||
| 185 | GALNT7 | 13493 | -1.029 | -0.0512 | No | ||
| 186 | MTMR3 | 13708 | -1.077 | -0.0597 | No | ||
| 187 | SOCS4 | 13892 | -1.122 | -0.0663 | No | ||
| 188 | TEF | 13964 | -1.141 | -0.0668 | No | ||
| 189 | WARS | 13972 | -1.141 | -0.0638 | No | ||
| 190 | SUV39H1 | 14153 | -1.184 | -0.0701 | No | ||
| 191 | DIRAS1 | 14214 | -1.199 | -0.0698 | No | ||
| 192 | PPP1CA | 14222 | -1.201 | -0.0666 | No | ||
| 193 | DNAJB5 | 14269 | -1.211 | -0.0656 | No | ||
| 194 | HCN4 | 14354 | -1.233 | -0.0665 | No | ||
| 195 | ATP1B4 | 14370 | -1.237 | -0.0636 | No | ||
| 196 | MAP6 | 14409 | -1.249 | -0.0620 | No | ||
| 197 | MADD | 14631 | -1.298 | -0.0702 | No | ||
| 198 | SLC27A4 | 14645 | -1.302 | -0.0670 | No | ||
| 199 | MFN1 | 14664 | -1.305 | -0.0641 | No | ||
| 200 | LOXL1 | 14882 | -1.356 | -0.0719 | No | ||
| 201 | BMPR2 | 14947 | -1.375 | -0.0713 | No | ||
| 202 | MASP1 | 15069 | -1.404 | -0.0738 | No | ||
| 203 | PTPRF | 15100 | -1.412 | -0.0712 | No | ||
| 204 | ZSWIM4 | 15104 | -1.412 | -0.0672 | No | ||
| 205 | VDR | 15254 | -1.452 | -0.0710 | No | ||
| 206 | HOXC6 | 15488 | -1.529 | -0.0791 | No | ||
| 207 | PHF15 | 15535 | -1.543 | -0.0771 | No | ||
| 208 | ATOH8 | 15621 | -1.570 | -0.0770 | No | ||
| 209 | SAMD10 | 15859 | -1.648 | -0.0851 | No | ||
| 210 | ANKRD50 | 15995 | -1.694 | -0.0874 | No | ||
| 211 | COPS7B | 16037 | -1.711 | -0.0846 | No | ||
| 212 | ADAM11 | 16172 | -1.757 | -0.0866 | No | ||
| 213 | MAMDC2 | 16180 | -1.759 | -0.0818 | No | ||
| 214 | OAZ2 | 16520 | -1.882 | -0.0947 | No | ||
| 215 | RNF121 | 16619 | -1.927 | -0.0943 | No | ||
| 216 | EIF4EBP1 | 16641 | -1.937 | -0.0897 | No | ||
| 217 | ASB6 | 16903 | -2.059 | -0.0978 | No | ||
| 218 | SET | 16952 | -2.082 | -0.0942 | No | ||
| 219 | UBE2G1 | 17068 | -2.144 | -0.0942 | No | ||
| 220 | PLXNA1 | 17082 | -2.152 | -0.0885 | No | ||
| 221 | DYNLT3 | 17118 | -2.177 | -0.0839 | No | ||
| 222 | SLC7A6 | 17197 | -2.227 | -0.0816 | No | ||
| 223 | SERTAD3 | 17337 | -2.317 | -0.0823 | No | ||
| 224 | SNAI1 | 17371 | -2.345 | -0.0771 | No | ||
| 225 | HIVEP2 | 17465 | -2.405 | -0.0751 | No | ||
| 226 | GGTL3 | 17550 | -2.455 | -0.0724 | No | ||
| 227 | MTUS1 | 17655 | -2.556 | -0.0705 | No | ||
| 228 | TRPS1 | 17732 | -2.624 | -0.0668 | No | ||
| 229 | GRSF1 | 17778 | -2.657 | -0.0614 | No | ||
| 230 | RFXANK | 17820 | -2.693 | -0.0556 | No | ||
| 231 | DOCK3 | 17921 | -2.791 | -0.0528 | No | ||
| 232 | M6PR | 17976 | -2.872 | -0.0472 | No | ||
| 233 | STARD13 | 18084 | -3.010 | -0.0442 | No | ||
| 234 | FLOT2 | 18110 | -3.043 | -0.0365 | No | ||
| 235 | NIPA1 | 18203 | -3.181 | -0.0321 | No | ||
| 236 | SLC25A15 | 18388 | -3.611 | -0.0314 | No | ||
| 237 | ITGA9 | 18512 | -4.204 | -0.0256 | No | ||
| 238 | IRF4 | 18590 | -5.255 | -0.0143 | No | ||
| 239 | ORC2L | 18591 | -5.271 | 0.0014 | No |