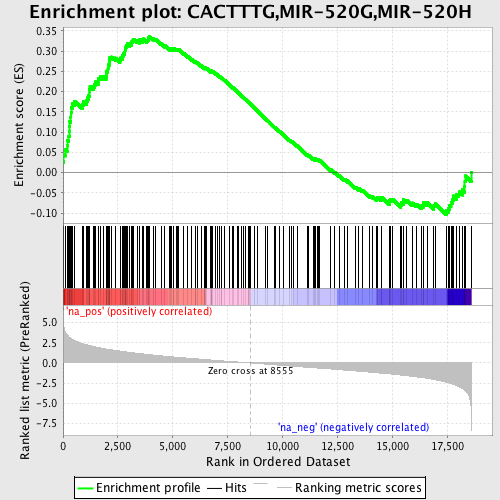
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CACTTTG,MIR-520G,MIR-520H |
| Enrichment Score (ES) | 0.33671725 |
| Normalized Enrichment Score (NES) | 1.7088449 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.06335129 |
| FWER p-Value | 0.388 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TCERG1 | 0 | 6.442 | 0.0266 | Yes | ||
| 2 | CENTD1 | 28 | 4.626 | 0.0443 | Yes | ||
| 3 | CIC | 90 | 3.897 | 0.0571 | Yes | ||
| 4 | MAP3K14 | 182 | 3.508 | 0.0667 | Yes | ||
| 5 | NCOA3 | 202 | 3.427 | 0.0798 | Yes | ||
| 6 | TGOLN2 | 259 | 3.233 | 0.0902 | Yes | ||
| 7 | NIN | 285 | 3.177 | 0.1019 | Yes | ||
| 8 | TNRC6A | 291 | 3.169 | 0.1148 | Yes | ||
| 9 | ZBTB7A | 304 | 3.137 | 0.1271 | Yes | ||
| 10 | PLAGL2 | 353 | 3.031 | 0.1370 | Yes | ||
| 11 | BRD1 | 359 | 3.020 | 0.1493 | Yes | ||
| 12 | PITX1 | 369 | 3.003 | 0.1612 | Yes | ||
| 13 | USP9X | 415 | 2.935 | 0.1709 | Yes | ||
| 14 | DMTF1 | 528 | 2.792 | 0.1763 | Yes | ||
| 15 | DDX11 | 867 | 2.424 | 0.1680 | Yes | ||
| 16 | MACF1 | 927 | 2.369 | 0.1746 | Yes | ||
| 17 | DNM2 | 1058 | 2.282 | 0.1770 | Yes | ||
| 18 | SS18L1 | 1095 | 2.242 | 0.1843 | Yes | ||
| 19 | E2F1 | 1158 | 2.188 | 0.1900 | Yes | ||
| 20 | TBL1X | 1203 | 2.156 | 0.1966 | Yes | ||
| 21 | CUL3 | 1206 | 2.154 | 0.2054 | Yes | ||
| 22 | EBF3 | 1224 | 2.136 | 0.2133 | Yes | ||
| 23 | ZDHHC9 | 1385 | 2.028 | 0.2130 | Yes | ||
| 24 | AHCTF1 | 1435 | 2.002 | 0.2186 | Yes | ||
| 25 | MAT2A | 1490 | 1.977 | 0.2239 | Yes | ||
| 26 | SENP1 | 1596 | 1.913 | 0.2261 | Yes | ||
| 27 | DMD | 1614 | 1.903 | 0.2330 | Yes | ||
| 28 | TLE4 | 1692 | 1.857 | 0.2365 | Yes | ||
| 29 | MARK4 | 1821 | 1.798 | 0.2370 | Yes | ||
| 30 | CHD9 | 1958 | 1.729 | 0.2368 | Yes | ||
| 31 | SKI | 1975 | 1.720 | 0.2430 | Yes | ||
| 32 | SSFA2 | 1980 | 1.719 | 0.2499 | Yes | ||
| 33 | EPHB2 | 2043 | 1.695 | 0.2536 | Yes | ||
| 34 | DYRK1A | 2046 | 1.693 | 0.2605 | Yes | ||
| 35 | ETS1 | 2076 | 1.684 | 0.2659 | Yes | ||
| 36 | PFN2 | 2097 | 1.676 | 0.2717 | Yes | ||
| 37 | CSMD3 | 2108 | 1.668 | 0.2781 | Yes | ||
| 38 | CFL2 | 2133 | 1.658 | 0.2836 | Yes | ||
| 39 | ZFAND3 | 2227 | 1.616 | 0.2853 | Yes | ||
| 40 | MAPK9 | 2391 | 1.543 | 0.2828 | Yes | ||
| 41 | POU4F2 | 2593 | 1.468 | 0.2780 | Yes | ||
| 42 | HMGB3 | 2616 | 1.460 | 0.2828 | Yes | ||
| 43 | SALL1 | 2691 | 1.431 | 0.2847 | Yes | ||
| 44 | SORL1 | 2718 | 1.423 | 0.2892 | Yes | ||
| 45 | MKNK1 | 2750 | 1.409 | 0.2933 | Yes | ||
| 46 | USP12 | 2793 | 1.393 | 0.2968 | Yes | ||
| 47 | FASTK | 2820 | 1.380 | 0.3011 | Yes | ||
| 48 | FLRT3 | 2827 | 1.379 | 0.3065 | Yes | ||
| 49 | DNAJB6 | 2864 | 1.366 | 0.3102 | Yes | ||
| 50 | CCND2 | 2900 | 1.351 | 0.3139 | Yes | ||
| 51 | PPP3CA | 2926 | 1.342 | 0.3181 | Yes | ||
| 52 | PRICKLE2 | 3039 | 1.308 | 0.3174 | Yes | ||
| 53 | ARMC8 | 3124 | 1.283 | 0.3182 | Yes | ||
| 54 | PCDHA10 | 3135 | 1.279 | 0.3229 | Yes | ||
| 55 | THRA | 3180 | 1.263 | 0.3257 | Yes | ||
| 56 | PLEKHM1 | 3211 | 1.250 | 0.3293 | Yes | ||
| 57 | NHLH1 | 3367 | 1.201 | 0.3258 | Yes | ||
| 58 | MAP3K9 | 3485 | 1.163 | 0.3243 | Yes | ||
| 59 | CHUK | 3488 | 1.162 | 0.3290 | Yes | ||
| 60 | TRIM33 | 3626 | 1.116 | 0.3262 | Yes | ||
| 61 | PCDHA7 | 3645 | 1.111 | 0.3298 | Yes | ||
| 62 | ZBTB1 | 3806 | 1.056 | 0.3255 | Yes | ||
| 63 | SUMF1 | 3859 | 1.040 | 0.3270 | Yes | ||
| 64 | RUNX2 | 3878 | 1.034 | 0.3303 | Yes | ||
| 65 | SLC6A9 | 3894 | 1.029 | 0.3337 | Yes | ||
| 66 | CASP8 | 3918 | 1.024 | 0.3367 | Yes | ||
| 67 | TNFAIP1 | 4098 | 0.977 | 0.3310 | No | ||
| 68 | TNKS1BP1 | 4208 | 0.946 | 0.3291 | No | ||
| 69 | PTK2B | 4470 | 0.877 | 0.3185 | No | ||
| 70 | CBX2 | 4635 | 0.839 | 0.3131 | No | ||
| 71 | TNFSF11 | 4842 | 0.785 | 0.3052 | No | ||
| 72 | FNDC3A | 4900 | 0.772 | 0.3053 | No | ||
| 73 | ARID4B | 4955 | 0.756 | 0.3055 | No | ||
| 74 | CNR1 | 5033 | 0.741 | 0.3044 | No | ||
| 75 | PLCB1 | 5051 | 0.735 | 0.3065 | No | ||
| 76 | EFTUD1 | 5167 | 0.709 | 0.3032 | No | ||
| 77 | GRM7 | 5192 | 0.705 | 0.3048 | No | ||
| 78 | MYCBP | 5274 | 0.683 | 0.3032 | No | ||
| 79 | BAI2 | 5498 | 0.630 | 0.2937 | No | ||
| 80 | WDR1 | 5688 | 0.594 | 0.2860 | No | ||
| 81 | ABCA1 | 5862 | 0.556 | 0.2789 | No | ||
| 82 | SLC39A14 | 6018 | 0.520 | 0.2726 | No | ||
| 83 | FOXO1A | 6042 | 0.516 | 0.2735 | No | ||
| 84 | APBB2 | 6106 | 0.499 | 0.2721 | No | ||
| 85 | TFEB | 6297 | 0.460 | 0.2637 | No | ||
| 86 | GOLGA7 | 6449 | 0.423 | 0.2573 | No | ||
| 87 | ARL4C | 6463 | 0.421 | 0.2583 | No | ||
| 88 | PAFAH1B2 | 6484 | 0.415 | 0.2590 | No | ||
| 89 | SMAD7 | 6544 | 0.403 | 0.2574 | No | ||
| 90 | NLK | 6725 | 0.363 | 0.2492 | No | ||
| 91 | TRPC5 | 6743 | 0.359 | 0.2497 | No | ||
| 92 | EPHA5 | 6749 | 0.359 | 0.2510 | No | ||
| 93 | LACE1 | 6782 | 0.352 | 0.2507 | No | ||
| 94 | KLF12 | 6826 | 0.342 | 0.2498 | No | ||
| 95 | YPEL2 | 6963 | 0.311 | 0.2437 | No | ||
| 96 | HIF1A | 7018 | 0.299 | 0.2420 | No | ||
| 97 | PLOD2 | 7145 | 0.272 | 0.2363 | No | ||
| 98 | MIPOL1 | 7207 | 0.260 | 0.2340 | No | ||
| 99 | KHDRBS1 | 7221 | 0.258 | 0.2344 | No | ||
| 100 | PRKAR2A | 7362 | 0.230 | 0.2278 | No | ||
| 101 | DBN1 | 7580 | 0.186 | 0.2168 | No | ||
| 102 | MEF2D | 7737 | 0.159 | 0.2090 | No | ||
| 103 | MGEA5 | 7748 | 0.157 | 0.2091 | No | ||
| 104 | MAPRE1 | 7944 | 0.117 | 0.1990 | No | ||
| 105 | SLC30A3 | 7975 | 0.113 | 0.1978 | No | ||
| 106 | EPAS1 | 8111 | 0.089 | 0.1909 | No | ||
| 107 | RHO | 8205 | 0.073 | 0.1861 | No | ||
| 108 | WDR42A | 8331 | 0.048 | 0.1795 | No | ||
| 109 | LIMK1 | 8436 | 0.023 | 0.1740 | No | ||
| 110 | PCDHA3 | 8457 | 0.018 | 0.1730 | No | ||
| 111 | FAM60A | 8475 | 0.015 | 0.1721 | No | ||
| 112 | SATB2 | 8555 | -0.000 | 0.1679 | No | ||
| 113 | DAPK2 | 8742 | -0.039 | 0.1579 | No | ||
| 114 | PCDHA6 | 8845 | -0.059 | 0.1526 | No | ||
| 115 | EIF4G2 | 9229 | -0.142 | 0.1325 | No | ||
| 116 | ATP2B2 | 9317 | -0.163 | 0.1284 | No | ||
| 117 | FURIN | 9633 | -0.227 | 0.1123 | No | ||
| 118 | ATXN1 | 9674 | -0.236 | 0.1111 | No | ||
| 119 | JUB | 9857 | -0.276 | 0.1024 | No | ||
| 120 | PCDHA5 | 10063 | -0.324 | 0.0926 | No | ||
| 121 | PAPOLG | 10317 | -0.379 | 0.0804 | No | ||
| 122 | SEMA4C | 10410 | -0.396 | 0.0771 | No | ||
| 123 | NPAS2 | 10490 | -0.410 | 0.0745 | No | ||
| 124 | SPRY4 | 10667 | -0.447 | 0.0668 | No | ||
| 125 | CCNJ | 11129 | -0.540 | 0.0440 | No | ||
| 126 | FBN2 | 11206 | -0.555 | 0.0422 | No | ||
| 127 | SLC1A2 | 11394 | -0.593 | 0.0345 | No | ||
| 128 | CDKN1A | 11472 | -0.609 | 0.0328 | No | ||
| 129 | RHOV | 11481 | -0.611 | 0.0349 | No | ||
| 130 | USP37 | 11579 | -0.628 | 0.0323 | No | ||
| 131 | KPNA3 | 11646 | -0.642 | 0.0314 | No | ||
| 132 | SMAD6 | 11706 | -0.651 | 0.0308 | No | ||
| 133 | PCYT1B | 12205 | -0.749 | 0.0069 | No | ||
| 134 | SMOC2 | 12391 | -0.789 | 0.0002 | No | ||
| 135 | SLC2A3 | 12577 | -0.831 | -0.0064 | No | ||
| 136 | ENC1 | 12817 | -0.883 | -0.0157 | No | ||
| 137 | FLRT2 | 12966 | -0.913 | -0.0200 | No | ||
| 138 | SOCS6 | 13337 | -0.995 | -0.0359 | No | ||
| 139 | RUNX1 | 13479 | -1.027 | -0.0393 | No | ||
| 140 | TBX3 | 13629 | -1.061 | -0.0430 | No | ||
| 141 | CELSR3 | 13978 | -1.143 | -0.0572 | No | ||
| 142 | NPAS3 | 14082 | -1.165 | -0.0579 | No | ||
| 143 | DNAJB5 | 14269 | -1.211 | -0.0630 | No | ||
| 144 | EFNB1 | 14313 | -1.224 | -0.0603 | No | ||
| 145 | LHX8 | 14492 | -1.268 | -0.0647 | No | ||
| 146 | PRRX1 | 14518 | -1.273 | -0.0608 | No | ||
| 147 | PCDHA12 | 14867 | -1.354 | -0.0741 | No | ||
| 148 | SPRED1 | 14874 | -1.355 | -0.0688 | No | ||
| 149 | ETF1 | 14934 | -1.370 | -0.0663 | No | ||
| 150 | KPNA1 | 15022 | -1.395 | -0.0653 | No | ||
| 151 | SUHW2 | 15399 | -1.501 | -0.0795 | No | ||
| 152 | NR4A3 | 15420 | -1.508 | -0.0743 | No | ||
| 153 | RFX4 | 15500 | -1.533 | -0.0722 | No | ||
| 154 | RABGAP1 | 15511 | -1.536 | -0.0664 | No | ||
| 155 | PCDHA1 | 15667 | -1.584 | -0.0683 | No | ||
| 156 | SERINC1 | 15917 | -1.668 | -0.0749 | No | ||
| 157 | ABHD2 | 16098 | -1.733 | -0.0775 | No | ||
| 158 | TNFSF12 | 16323 | -1.807 | -0.0822 | No | ||
| 159 | TTN | 16405 | -1.833 | -0.0790 | No | ||
| 160 | VPS26A | 16428 | -1.844 | -0.0725 | No | ||
| 161 | PCDHA11 | 16588 | -1.911 | -0.0733 | No | ||
| 162 | CELSR2 | 16902 | -2.059 | -0.0817 | No | ||
| 163 | SET | 16952 | -2.082 | -0.0758 | No | ||
| 164 | HBXIP | 17460 | -2.405 | -0.0933 | No | ||
| 165 | PTPN13 | 17556 | -2.459 | -0.0883 | No | ||
| 166 | MAF1 | 17601 | -2.509 | -0.0803 | No | ||
| 167 | VLDLR | 17680 | -2.580 | -0.0739 | No | ||
| 168 | TRPS1 | 17732 | -2.624 | -0.0658 | No | ||
| 169 | PPP6C | 17785 | -2.665 | -0.0576 | No | ||
| 170 | EIF4E | 17937 | -2.816 | -0.0541 | No | ||
| 171 | CALU | 18043 | -2.955 | -0.0476 | No | ||
| 172 | AZIN1 | 18187 | -3.157 | -0.0423 | No | ||
| 173 | DPYSL5 | 18298 | -3.368 | -0.0343 | No | ||
| 174 | DHDDS | 18316 | -3.412 | -0.0212 | No | ||
| 175 | NEUROG2 | 18343 | -3.483 | -0.0082 | No | ||
| 176 | NR4A2 | 18597 | -5.541 | 0.0010 | No |
