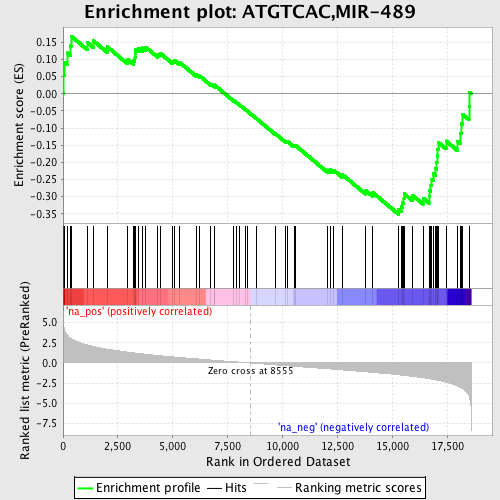
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ATGTCAC,MIR-489 |
| Enrichment Score (ES) | -0.35205275 |
| Normalized Enrichment Score (NES) | -1.4291291 |
| Nominal p-value | 0.029363785 |
| FDR q-value | 0.521093 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ATP2A3 | 6 | 5.724 | 0.0550 | No | ||
| 2 | BCL11A | 59 | 4.158 | 0.0924 | No | ||
| 3 | NSD1 | 186 | 3.492 | 0.1193 | No | ||
| 4 | RBBP6 | 352 | 3.032 | 0.1398 | No | ||
| 5 | RERE | 362 | 3.014 | 0.1684 | No | ||
| 6 | MDM1 | 1112 | 2.225 | 0.1495 | No | ||
| 7 | CLOCK | 1378 | 2.031 | 0.1549 | No | ||
| 8 | WNT5A | 2012 | 1.708 | 0.1372 | No | ||
| 9 | GPD1 | 2935 | 1.339 | 0.1005 | No | ||
| 10 | BACH2 | 3218 | 1.249 | 0.0973 | No | ||
| 11 | ATRX | 3275 | 1.228 | 0.1062 | No | ||
| 12 | BAALC | 3296 | 1.222 | 0.1169 | No | ||
| 13 | BMP7 | 3308 | 1.220 | 0.1281 | No | ||
| 14 | DDX3Y | 3420 | 1.185 | 0.1336 | No | ||
| 15 | CUGBP1 | 3616 | 1.119 | 0.1339 | No | ||
| 16 | ASXL2 | 3760 | 1.074 | 0.1366 | No | ||
| 17 | NCOA1 | 4311 | 0.923 | 0.1158 | No | ||
| 18 | BTG1 | 4415 | 0.893 | 0.1189 | No | ||
| 19 | PARP12 | 4984 | 0.751 | 0.0956 | No | ||
| 20 | PDE4D | 5085 | 0.727 | 0.0972 | No | ||
| 21 | SMAD3 | 5295 | 0.677 | 0.0925 | No | ||
| 22 | OPN3 | 6056 | 0.512 | 0.0565 | No | ||
| 23 | RBM12 | 6221 | 0.476 | 0.0522 | No | ||
| 24 | NLK | 6725 | 0.363 | 0.0286 | No | ||
| 25 | DDX3X | 6889 | 0.328 | 0.0230 | No | ||
| 26 | ARHGEF5 | 6894 | 0.327 | 0.0259 | No | ||
| 27 | UNC5D | 7779 | 0.152 | -0.0203 | No | ||
| 28 | POU3F2 | 7898 | 0.128 | -0.0254 | No | ||
| 29 | TRIB2 | 8056 | 0.099 | -0.0329 | No | ||
| 30 | RARB | 8308 | 0.054 | -0.0459 | No | ||
| 31 | BCL11B | 8393 | 0.035 | -0.0501 | No | ||
| 32 | NRG1 | 8797 | -0.049 | -0.0713 | No | ||
| 33 | SP4 | 9686 | -0.238 | -0.1169 | No | ||
| 34 | ATP2B3 | 10155 | -0.340 | -0.1389 | No | ||
| 35 | FBN1 | 10232 | -0.359 | -0.1395 | No | ||
| 36 | HUNK | 10523 | -0.416 | -0.1511 | No | ||
| 37 | PURG | 10599 | -0.433 | -0.1509 | No | ||
| 38 | EIF4A1 | 12050 | -0.714 | -0.2222 | No | ||
| 39 | XPO1 | 12176 | -0.742 | -0.2218 | No | ||
| 40 | PPP1CB | 12344 | -0.780 | -0.2232 | No | ||
| 41 | ARID5A | 12733 | -0.867 | -0.2358 | No | ||
| 42 | ZADH2 | 13784 | -1.094 | -0.2818 | No | ||
| 43 | CXXC5 | 14121 | -1.176 | -0.2885 | No | ||
| 44 | SGK | 15300 | -1.466 | -0.3379 | Yes | ||
| 45 | NR4A3 | 15420 | -1.508 | -0.3297 | Yes | ||
| 46 | SOX30 | 15481 | -1.528 | -0.3182 | Yes | ||
| 47 | PHF15 | 15535 | -1.543 | -0.3061 | Yes | ||
| 48 | THADA | 15539 | -1.544 | -0.2914 | Yes | ||
| 49 | BRSK2 | 15933 | -1.673 | -0.2964 | Yes | ||
| 50 | TTN | 16405 | -1.833 | -0.3041 | Yes | ||
| 51 | SNX3 | 16679 | -1.954 | -0.2999 | Yes | ||
| 52 | STC2 | 16686 | -1.959 | -0.2813 | Yes | ||
| 53 | RIOK3 | 16764 | -1.988 | -0.2662 | Yes | ||
| 54 | DCX | 16804 | -2.008 | -0.2489 | Yes | ||
| 55 | RSU1 | 16885 | -2.052 | -0.2334 | Yes | ||
| 56 | NTRK3 | 16980 | -2.096 | -0.2182 | Yes | ||
| 57 | ADAMTS5 | 17015 | -2.113 | -0.1996 | Yes | ||
| 58 | RANBP5 | 17041 | -2.128 | -0.1804 | Yes | ||
| 59 | LIN7C | 17069 | -2.145 | -0.1611 | Yes | ||
| 60 | SCN3A | 17112 | -2.174 | -0.1424 | Yes | ||
| 61 | ST8SIA2 | 17456 | -2.404 | -0.1376 | Yes | ||
| 62 | PCDH10 | 17988 | -2.886 | -0.1384 | Yes | ||
| 63 | SOX12 | 18120 | -3.060 | -0.1158 | Yes | ||
| 64 | OXR1 | 18146 | -3.102 | -0.0872 | Yes | ||
| 65 | SRGAP2 | 18193 | -3.169 | -0.0591 | Yes | ||
| 66 | FAF1 | 18501 | -4.134 | -0.0357 | Yes | ||
| 67 | UBXD2 | 18526 | -4.326 | 0.0049 | Yes |
