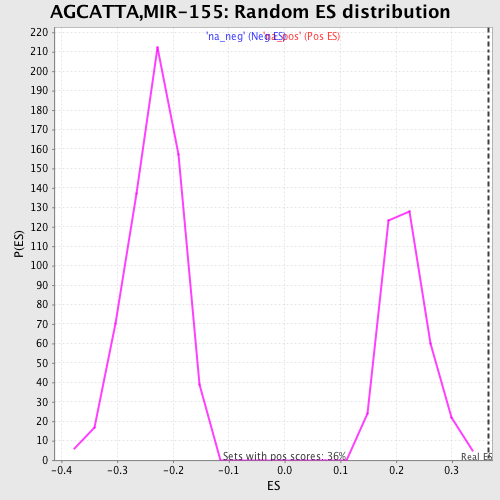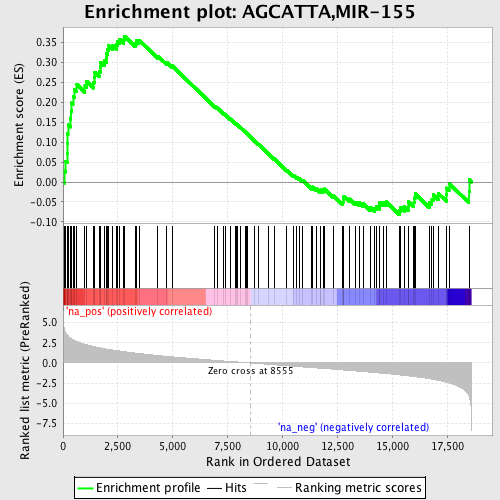
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | AGCATTA,MIR-155 |
| Enrichment Score (ES) | 0.36614797 |
| Normalized Enrichment Score (NES) | 1.6771111 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.06075123 |
| FWER p-Value | 0.47 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | STRN3 | 46 | 4.330 | 0.0280 | Yes | ||
| 2 | CHD7 | 100 | 3.875 | 0.0525 | Yes | ||
| 3 | MAP3K14 | 182 | 3.508 | 0.0729 | Yes | ||
| 4 | MAP3K7IP2 | 188 | 3.487 | 0.0972 | Yes | ||
| 5 | SYPL1 | 198 | 3.439 | 0.1209 | Yes | ||
| 6 | CUTL1 | 223 | 3.360 | 0.1433 | Yes | ||
| 7 | CUGBP2 | 320 | 3.105 | 0.1600 | Yes | ||
| 8 | BRD1 | 359 | 3.020 | 0.1793 | Yes | ||
| 9 | MECP2 | 392 | 2.971 | 0.1985 | Yes | ||
| 10 | NFAT5 | 451 | 2.890 | 0.2157 | Yes | ||
| 11 | UPP2 | 506 | 2.819 | 0.2327 | Yes | ||
| 12 | TOMM20 | 626 | 2.667 | 0.2451 | Yes | ||
| 13 | CSNK1G2 | 995 | 2.323 | 0.2416 | Yes | ||
| 14 | CD47 | 1071 | 2.267 | 0.2535 | Yes | ||
| 15 | CNTN4 | 1398 | 2.020 | 0.2501 | Yes | ||
| 16 | NFIX | 1429 | 2.005 | 0.2626 | Yes | ||
| 17 | PICALM | 1442 | 1.999 | 0.2761 | Yes | ||
| 18 | SMARCA4 | 1648 | 1.884 | 0.2783 | Yes | ||
| 19 | TLE4 | 1692 | 1.857 | 0.2891 | Yes | ||
| 20 | DNAJB1 | 1710 | 1.847 | 0.3012 | Yes | ||
| 21 | PTPN2 | 1887 | 1.766 | 0.3041 | Yes | ||
| 22 | SKI | 1975 | 1.720 | 0.3116 | Yes | ||
| 23 | GDF6 | 1985 | 1.717 | 0.3232 | Yes | ||
| 24 | PSIP1 | 2036 | 1.697 | 0.3324 | Yes | ||
| 25 | ETS1 | 2076 | 1.684 | 0.3422 | Yes | ||
| 26 | ARID2 | 2269 | 1.597 | 0.3431 | Yes | ||
| 27 | PCDH9 | 2448 | 1.522 | 0.3442 | Yes | ||
| 28 | FBXO33 | 2482 | 1.511 | 0.3531 | Yes | ||
| 29 | SGK3 | 2575 | 1.476 | 0.3585 | Yes | ||
| 30 | SATB1 | 2773 | 1.400 | 0.3578 | Yes | ||
| 31 | MIDN | 2800 | 1.389 | 0.3661 | Yes | ||
| 32 | DYNC1I1 | 3302 | 1.221 | 0.3477 | No | ||
| 33 | CAB39 | 3334 | 1.210 | 0.3546 | No | ||
| 34 | CEBPB | 3458 | 1.173 | 0.3562 | No | ||
| 35 | INPP5D | 4321 | 0.920 | 0.3161 | No | ||
| 36 | ZIC3 | 4729 | 0.813 | 0.2999 | No | ||
| 37 | SLA | 4963 | 0.755 | 0.2926 | No | ||
| 38 | NDFIP1 | 6917 | 0.321 | 0.1894 | No | ||
| 39 | HIF1A | 7018 | 0.299 | 0.1862 | No | ||
| 40 | BACH1 | 7328 | 0.236 | 0.1711 | No | ||
| 41 | BCORL1 | 7403 | 0.219 | 0.1687 | No | ||
| 42 | ACTA1 | 7619 | 0.181 | 0.1584 | No | ||
| 43 | EDG1 | 7848 | 0.137 | 0.1470 | No | ||
| 44 | ITK | 7885 | 0.130 | 0.1460 | No | ||
| 45 | SGCZ | 7959 | 0.114 | 0.1428 | No | ||
| 46 | ACVR2A | 8074 | 0.095 | 0.1374 | No | ||
| 47 | RAB11FIP2 | 8295 | 0.058 | 0.1259 | No | ||
| 48 | FGF7 | 8345 | 0.045 | 0.1236 | No | ||
| 49 | MEIS1 | 8414 | 0.030 | 0.1201 | No | ||
| 50 | H3F3A | 8716 | -0.034 | 0.1041 | No | ||
| 51 | MYO10 | 8904 | -0.071 | 0.0945 | No | ||
| 52 | ELL2 | 9371 | -0.171 | 0.0705 | No | ||
| 53 | SPI1 | 9616 | -0.224 | 0.0589 | No | ||
| 54 | IKBKE | 10197 | -0.350 | 0.0301 | No | ||
| 55 | SOCS1 | 10482 | -0.409 | 0.0176 | No | ||
| 56 | KRAS | 10637 | -0.440 | 0.0124 | No | ||
| 57 | SERAC1 | 10751 | -0.465 | 0.0096 | No | ||
| 58 | SOX11 | 10920 | -0.499 | 0.0040 | No | ||
| 59 | TRIM2 | 11333 | -0.581 | -0.0141 | No | ||
| 60 | MYB | 11377 | -0.589 | -0.0123 | No | ||
| 61 | YWHAE | 11544 | -0.621 | -0.0169 | No | ||
| 62 | WEE1 | 11720 | -0.655 | -0.0217 | No | ||
| 63 | SLC39A10 | 11752 | -0.660 | -0.0187 | No | ||
| 64 | CTLA4 | 11871 | -0.685 | -0.0203 | No | ||
| 65 | MYO1D | 11907 | -0.690 | -0.0173 | No | ||
| 66 | SCG2 | 12321 | -0.776 | -0.0341 | No | ||
| 67 | SP3 | 12743 | -0.869 | -0.0507 | No | ||
| 68 | BOC | 12760 | -0.872 | -0.0455 | No | ||
| 69 | OGN | 12761 | -0.872 | -0.0393 | No | ||
| 70 | DNAJB7 | 12800 | -0.879 | -0.0352 | No | ||
| 71 | MAP3K10 | 13057 | -0.932 | -0.0424 | No | ||
| 72 | KCNN3 | 13334 | -0.995 | -0.0503 | No | ||
| 73 | CDC73 | 13488 | -1.028 | -0.0513 | No | ||
| 74 | BCAT1 | 13684 | -1.072 | -0.0543 | No | ||
| 75 | LRP1B | 14012 | -1.151 | -0.0638 | No | ||
| 76 | QKI | 14200 | -1.196 | -0.0655 | No | ||
| 77 | SOX1 | 14271 | -1.212 | -0.0607 | No | ||
| 78 | BCAP29 | 14403 | -1.246 | -0.0590 | No | ||
| 79 | ARL5B | 14435 | -1.255 | -0.0518 | No | ||
| 80 | RAB6A | 14587 | -1.289 | -0.0509 | No | ||
| 81 | RPS6KA3 | 14730 | -1.321 | -0.0493 | No | ||
| 82 | TIMM8A | 15325 | -1.474 | -0.0709 | No | ||
| 83 | SSH2 | 15380 | -1.495 | -0.0633 | No | ||
| 84 | SDCBP | 15557 | -1.549 | -0.0619 | No | ||
| 85 | CSF1R | 15741 | -1.608 | -0.0605 | No | ||
| 86 | RCN2 | 15747 | -1.609 | -0.0494 | No | ||
| 87 | SEMA5A | 15991 | -1.694 | -0.0506 | No | ||
| 88 | PKIA | 16018 | -1.704 | -0.0400 | No | ||
| 89 | MARCH7 | 16041 | -1.712 | -0.0291 | No | ||
| 90 | UBE2D3 | 16692 | -1.962 | -0.0503 | No | ||
| 91 | GPR85 | 16811 | -2.011 | -0.0425 | No | ||
| 92 | KBTBD2 | 16880 | -2.050 | -0.0318 | No | ||
| 93 | GPM6B | 17094 | -2.160 | -0.0280 | No | ||
| 94 | HIVEP2 | 17465 | -2.405 | -0.0311 | No | ||
| 95 | HBP1 | 17481 | -2.413 | -0.0149 | No | ||
| 96 | YWHAZ | 17591 | -2.498 | -0.0031 | No | ||
| 97 | RAB1A | 18499 | -4.108 | -0.0231 | No | ||
| 98 | CARHSP1 | 18508 | -4.171 | 0.0058 | No |