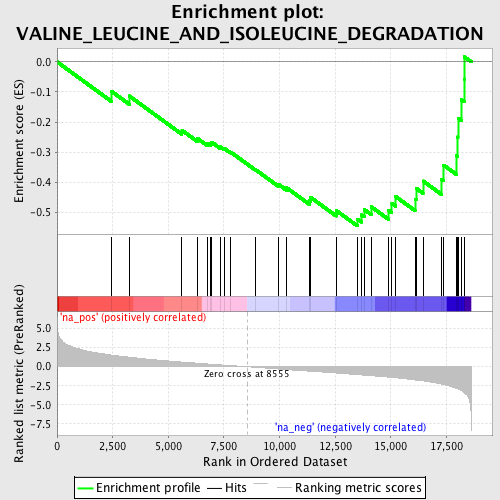
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION |
| Enrichment Score (ES) | -0.5452865 |
| Normalized Enrichment Score (NES) | -1.9015424 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.02404537 |
| FWER p-Value | 0.207 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ALDH2 | 2451 | 1.521 | -0.0982 | No | ||
| 2 | HADHA | 3240 | 1.241 | -0.1132 | No | ||
| 3 | MCCC1 | 5612 | 0.608 | -0.2273 | No | ||
| 4 | IVD | 6293 | 0.461 | -0.2537 | No | ||
| 5 | ALDH1A2 | 6767 | 0.355 | -0.2713 | No | ||
| 6 | ALDH3A1 | 6906 | 0.323 | -0.2716 | No | ||
| 7 | OXCT1 | 6956 | 0.312 | -0.2673 | No | ||
| 8 | PCCB | 7338 | 0.235 | -0.2826 | No | ||
| 9 | ALDH1A1 | 7505 | 0.199 | -0.2872 | No | ||
| 10 | MUT | 7797 | 0.148 | -0.2995 | No | ||
| 11 | ALDH9A1 | 8910 | -0.072 | -0.3578 | No | ||
| 12 | BCKDHB | 9958 | -0.298 | -0.4075 | No | ||
| 13 | ALDH1A3 | 10315 | -0.378 | -0.4183 | No | ||
| 14 | ACADM | 11357 | -0.585 | -0.4614 | No | ||
| 15 | HIBADH | 11382 | -0.590 | -0.4496 | No | ||
| 16 | ALDH6A1 | 12545 | -0.823 | -0.4939 | No | ||
| 17 | ACAT1 | 13500 | -1.030 | -0.5225 | Yes | ||
| 18 | BCAT1 | 13684 | -1.072 | -0.5086 | Yes | ||
| 19 | EHHADH | 13804 | -1.102 | -0.4906 | Yes | ||
| 20 | ACADL | 14125 | -1.177 | -0.4818 | Yes | ||
| 21 | ACAT2 | 14887 | -1.358 | -0.4927 | Yes | ||
| 22 | MCEE | 15049 | -1.400 | -0.4704 | Yes | ||
| 23 | AOX1 | 15224 | -1.443 | -0.4479 | Yes | ||
| 24 | ACADSB | 16090 | -1.731 | -0.4561 | Yes | ||
| 25 | ACAA2 | 16140 | -1.745 | -0.4202 | Yes | ||
| 26 | PCCA | 16449 | -1.849 | -0.3958 | Yes | ||
| 27 | ALDH1B1 | 17291 | -2.282 | -0.3906 | Yes | ||
| 28 | SDS | 17373 | -2.346 | -0.3431 | Yes | ||
| 29 | HADHB | 17943 | -2.825 | -0.3112 | Yes | ||
| 30 | ECHS1 | 17985 | -2.883 | -0.2496 | Yes | ||
| 31 | HMGCL | 18020 | -2.922 | -0.1868 | Yes | ||
| 32 | BCKDHA | 18168 | -3.133 | -0.1254 | Yes | ||
| 33 | ACADS | 18297 | -3.365 | -0.0578 | Yes | ||
| 34 | MCCC2 | 18305 | -3.387 | 0.0167 | Yes |