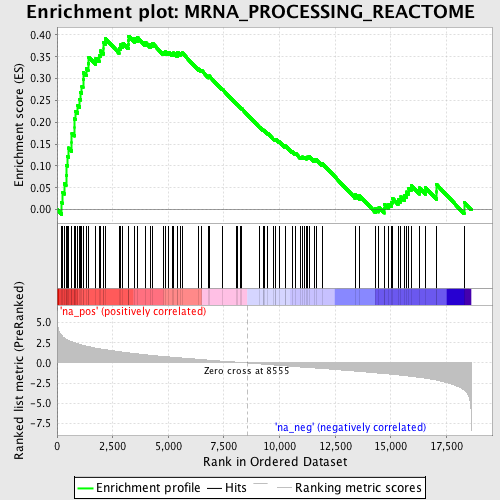
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | MRNA_PROCESSING_REACTOME |
| Enrichment Score (ES) | 0.39737523 |
| Normalized Enrichment Score (NES) | 1.8404924 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.053900443 |
| FWER p-Value | 0.484 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PAPOLA | 205 | 3.413 | 0.0160 | Yes | ||
| 2 | SFRS14 | 255 | 3.246 | 0.0391 | Yes | ||
| 3 | CUGBP2 | 320 | 3.105 | 0.0602 | Yes | ||
| 4 | RBM5 | 402 | 2.958 | 0.0793 | Yes | ||
| 5 | NXF1 | 413 | 2.935 | 0.1021 | Yes | ||
| 6 | DHX16 | 457 | 2.879 | 0.1226 | Yes | ||
| 7 | SFRS16 | 522 | 2.801 | 0.1413 | Yes | ||
| 8 | SFRS5 | 663 | 2.617 | 0.1545 | Yes | ||
| 9 | CLK4 | 666 | 2.615 | 0.1751 | Yes | ||
| 10 | DHX9 | 788 | 2.490 | 0.1883 | Yes | ||
| 11 | HNRPA3 | 790 | 2.487 | 0.2080 | Yes | ||
| 12 | SF3B2 | 832 | 2.453 | 0.2252 | Yes | ||
| 13 | SRRM1 | 930 | 2.365 | 0.2388 | Yes | ||
| 14 | CLK2 | 1018 | 2.306 | 0.2523 | Yes | ||
| 15 | PRPF8 | 1049 | 2.289 | 0.2689 | Yes | ||
| 16 | CD2BP2 | 1113 | 2.224 | 0.2831 | Yes | ||
| 17 | PABPN1 | 1166 | 2.180 | 0.2976 | Yes | ||
| 18 | SPOP | 1177 | 2.171 | 0.3143 | Yes | ||
| 19 | SNRPF | 1313 | 2.069 | 0.3234 | Yes | ||
| 20 | CPSF1 | 1393 | 2.024 | 0.3352 | Yes | ||
| 21 | SF3A1 | 1425 | 2.009 | 0.3494 | Yes | ||
| 22 | SNRPN | 1741 | 1.833 | 0.3469 | Yes | ||
| 23 | PTBP1 | 1890 | 1.765 | 0.3529 | Yes | ||
| 24 | SRPK1 | 1948 | 1.732 | 0.3636 | Yes | ||
| 25 | FUS | 2087 | 1.680 | 0.3695 | Yes | ||
| 26 | HNRPH1 | 2089 | 1.679 | 0.3827 | Yes | ||
| 27 | XRN2 | 2166 | 1.643 | 0.3916 | Yes | ||
| 28 | RNPS1 | 2784 | 1.395 | 0.3694 | Yes | ||
| 29 | HNRPA2B1 | 2838 | 1.375 | 0.3774 | Yes | ||
| 30 | DHX8 | 2959 | 1.332 | 0.3815 | Yes | ||
| 31 | DDX1 | 3200 | 1.252 | 0.3785 | Yes | ||
| 32 | HNRPH2 | 3212 | 1.250 | 0.3878 | Yes | ||
| 33 | CSTF3 | 3219 | 1.249 | 0.3974 | Yes | ||
| 34 | DHX38 | 3479 | 1.165 | 0.3926 | No | ||
| 35 | CUGBP1 | 3616 | 1.119 | 0.3942 | No | ||
| 36 | SUPT5H | 3964 | 1.011 | 0.3834 | No | ||
| 37 | DICER1 | 4210 | 0.946 | 0.3777 | No | ||
| 38 | SNRPD3 | 4291 | 0.929 | 0.3807 | No | ||
| 39 | HNRPU | 4777 | 0.801 | 0.3609 | No | ||
| 40 | SNRPB2 | 4880 | 0.775 | 0.3615 | No | ||
| 41 | RBM17 | 5007 | 0.746 | 0.3607 | No | ||
| 42 | SFRS8 | 5166 | 0.709 | 0.3577 | No | ||
| 43 | SNRP70 | 5245 | 0.691 | 0.3590 | No | ||
| 44 | DDIT3 | 5407 | 0.650 | 0.3555 | No | ||
| 45 | NONO | 5418 | 0.648 | 0.3601 | No | ||
| 46 | SFRS9 | 5536 | 0.624 | 0.3587 | No | ||
| 47 | SF3B4 | 5617 | 0.607 | 0.3592 | No | ||
| 48 | SF4 | 6366 | 0.442 | 0.3223 | No | ||
| 49 | U2AF1 | 6482 | 0.416 | 0.3194 | No | ||
| 50 | SFRS6 | 6815 | 0.345 | 0.3042 | No | ||
| 51 | PPM1G | 6827 | 0.342 | 0.3063 | No | ||
| 52 | PHF5A | 7423 | 0.215 | 0.2759 | No | ||
| 53 | NUDT21 | 8072 | 0.096 | 0.2417 | No | ||
| 54 | SF3A3 | 8098 | 0.091 | 0.2410 | No | ||
| 55 | CPSF2 | 8258 | 0.063 | 0.2330 | No | ||
| 56 | HNRPAB | 8309 | 0.054 | 0.2307 | No | ||
| 57 | DDX20 | 9097 | -0.111 | 0.1891 | No | ||
| 58 | CSTF2T | 9254 | -0.149 | 0.1818 | No | ||
| 59 | CPSF3 | 9265 | -0.152 | 0.1825 | No | ||
| 60 | POLR2A | 9330 | -0.165 | 0.1803 | No | ||
| 61 | HNRPL | 9468 | -0.192 | 0.1745 | No | ||
| 62 | DHX15 | 9741 | -0.251 | 0.1618 | No | ||
| 63 | SFRS7 | 9814 | -0.268 | 0.1600 | No | ||
| 64 | LSM2 | 9827 | -0.270 | 0.1615 | No | ||
| 65 | RNMT | 9985 | -0.304 | 0.1554 | No | ||
| 66 | SNRPA | 10246 | -0.362 | 0.1443 | No | ||
| 67 | HNRPD | 10263 | -0.367 | 0.1463 | No | ||
| 68 | SF3A2 | 10557 | -0.425 | 0.1339 | No | ||
| 69 | NCBP2 | 10705 | -0.453 | 0.1295 | No | ||
| 70 | DNAJC8 | 10947 | -0.505 | 0.1205 | No | ||
| 71 | CSTF2 | 11020 | -0.522 | 0.1208 | No | ||
| 72 | SNRPA1 | 11105 | -0.538 | 0.1205 | No | ||
| 73 | U2AF2 | 11193 | -0.552 | 0.1202 | No | ||
| 74 | COL2A1 | 11269 | -0.568 | 0.1206 | No | ||
| 75 | SF3B5 | 11335 | -0.581 | 0.1217 | No | ||
| 76 | SFRS10 | 11569 | -0.627 | 0.1141 | No | ||
| 77 | SNRPE | 11641 | -0.641 | 0.1154 | No | ||
| 78 | METTL3 | 11908 | -0.690 | 0.1065 | No | ||
| 79 | SRPK2 | 13395 | -1.008 | 0.0342 | No | ||
| 80 | SNRPD1 | 13591 | -1.052 | 0.0320 | No | ||
| 81 | PRPF4B | 14315 | -1.224 | 0.0027 | No | ||
| 82 | PRPF4 | 14466 | -1.260 | 0.0046 | No | ||
| 83 | HNRPC | 14696 | -1.313 | 0.0026 | No | ||
| 84 | SF3B1 | 14714 | -1.317 | 0.0122 | No | ||
| 85 | SNRPG | 14909 | -1.364 | 0.0125 | No | ||
| 86 | GIPC1 | 15027 | -1.396 | 0.0172 | No | ||
| 87 | SFRS2 | 15068 | -1.403 | 0.0262 | No | ||
| 88 | LSM7 | 15327 | -1.475 | 0.0240 | No | ||
| 89 | FUSIP1 | 15454 | -1.519 | 0.0292 | No | ||
| 90 | SNRPD2 | 15612 | -1.568 | 0.0332 | No | ||
| 91 | HNRPR | 15712 | -1.599 | 0.0405 | No | ||
| 92 | PRPF3 | 15808 | -1.628 | 0.0483 | No | ||
| 93 | CPSF4 | 15912 | -1.667 | 0.0559 | No | ||
| 94 | SNRPB | 16290 | -1.797 | 0.0498 | No | ||
| 95 | TXNL4A | 16565 | -1.900 | 0.0501 | No | ||
| 96 | CSTF1 | 17057 | -2.139 | 0.0405 | No | ||
| 97 | PTBP2 | 17061 | -2.142 | 0.0574 | No | ||
| 98 | RNGTT | 18295 | -3.349 | 0.0173 | No |