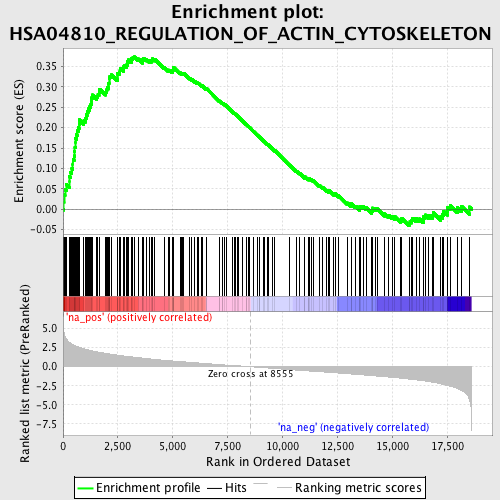
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HSA04810_REGULATION_OF_ACTIN_CYTOSKELETON |
| Enrichment Score (ES) | 0.37328893 |
| Normalized Enrichment Score (NES) | 1.8831712 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.042311583 |
| FWER p-Value | 0.347 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ARHGEF1 | 38 | 4.465 | 0.0167 | Yes | ||
| 2 | ITGB7 | 41 | 4.403 | 0.0352 | Yes | ||
| 3 | RAC2 | 88 | 3.901 | 0.0491 | Yes | ||
| 4 | WAS | 159 | 3.582 | 0.0604 | Yes | ||
| 5 | VCL | 279 | 3.193 | 0.0674 | Yes | ||
| 6 | SLC9A1 | 282 | 3.186 | 0.0807 | Yes | ||
| 7 | ACTN4 | 333 | 3.080 | 0.0909 | Yes | ||
| 8 | PAK4 | 401 | 2.958 | 0.0997 | Yes | ||
| 9 | PIP5K3 | 446 | 2.900 | 0.1095 | Yes | ||
| 10 | ROCK2 | 450 | 2.892 | 0.1216 | Yes | ||
| 11 | MAP2K1 | 502 | 2.825 | 0.1307 | Yes | ||
| 12 | FGFR1 | 529 | 2.788 | 0.1410 | Yes | ||
| 13 | PIP5K1C | 539 | 2.776 | 0.1522 | Yes | ||
| 14 | MSN | 562 | 2.740 | 0.1625 | Yes | ||
| 15 | CDC42 | 567 | 2.734 | 0.1738 | Yes | ||
| 16 | IQGAP1 | 617 | 2.684 | 0.1825 | Yes | ||
| 17 | MYH9 | 644 | 2.638 | 0.1922 | Yes | ||
| 18 | PPP1R12A | 694 | 2.591 | 0.2004 | Yes | ||
| 19 | LIMK2 | 731 | 2.551 | 0.2092 | Yes | ||
| 20 | ARPC1B | 745 | 2.527 | 0.2191 | Yes | ||
| 21 | RRAS | 951 | 2.351 | 0.2179 | Yes | ||
| 22 | CYFIP2 | 1033 | 2.300 | 0.2232 | Yes | ||
| 23 | ROCK1 | 1057 | 2.284 | 0.2316 | Yes | ||
| 24 | FGF14 | 1096 | 2.242 | 0.2389 | Yes | ||
| 25 | PIK3CG | 1139 | 2.200 | 0.2459 | Yes | ||
| 26 | ARPC2 | 1217 | 2.141 | 0.2508 | Yes | ||
| 27 | FGF5 | 1265 | 2.111 | 0.2571 | Yes | ||
| 28 | WASF2 | 1291 | 2.090 | 0.2645 | Yes | ||
| 29 | MAPK3 | 1303 | 2.079 | 0.2727 | Yes | ||
| 30 | WASL | 1329 | 2.059 | 0.2800 | Yes | ||
| 31 | FGF3 | 1539 | 1.944 | 0.2769 | Yes | ||
| 32 | APC | 1589 | 1.917 | 0.2823 | Yes | ||
| 33 | MAP2K2 | 1638 | 1.892 | 0.2876 | Yes | ||
| 34 | PDGFRB | 1659 | 1.880 | 0.2945 | Yes | ||
| 35 | ARHGEF6 | 1931 | 1.742 | 0.2871 | Yes | ||
| 36 | CSK | 1969 | 1.722 | 0.2923 | Yes | ||
| 37 | ARPC5L | 2001 | 1.711 | 0.2979 | Yes | ||
| 38 | VAV1 | 2063 | 1.688 | 0.3016 | Yes | ||
| 39 | SSH3 | 2068 | 1.686 | 0.3085 | Yes | ||
| 40 | FN1 | 2096 | 1.676 | 0.3141 | Yes | ||
| 41 | PFN2 | 2097 | 1.676 | 0.3212 | Yes | ||
| 42 | CFL2 | 2133 | 1.658 | 0.3263 | Yes | ||
| 43 | EGF | 2206 | 1.627 | 0.3292 | Yes | ||
| 44 | BDKRB2 | 2473 | 1.514 | 0.3211 | Yes | ||
| 45 | PTK2 | 2477 | 1.514 | 0.3274 | Yes | ||
| 46 | FGF8 | 2480 | 1.512 | 0.3336 | Yes | ||
| 47 | ITGB1 | 2557 | 1.485 | 0.3357 | Yes | ||
| 48 | FGF10 | 2591 | 1.469 | 0.3401 | Yes | ||
| 49 | SOS2 | 2609 | 1.463 | 0.3454 | Yes | ||
| 50 | GNG12 | 2745 | 1.412 | 0.3440 | Yes | ||
| 51 | RAF1 | 2767 | 1.403 | 0.3488 | Yes | ||
| 52 | FGF17 | 2810 | 1.384 | 0.3523 | Yes | ||
| 53 | SSH1 | 2894 | 1.353 | 0.3535 | Yes | ||
| 54 | ITGA10 | 2929 | 1.342 | 0.3573 | Yes | ||
| 55 | ITGAL | 2938 | 1.338 | 0.3625 | Yes | ||
| 56 | VAV2 | 2980 | 1.326 | 0.3659 | Yes | ||
| 57 | MYL7 | 3105 | 1.289 | 0.3646 | Yes | ||
| 58 | BDKRB1 | 3122 | 1.283 | 0.3691 | Yes | ||
| 59 | ITGB2 | 3183 | 1.262 | 0.3712 | Yes | ||
| 60 | FGFR2 | 3241 | 1.241 | 0.3733 | Yes | ||
| 61 | FGFR3 | 3428 | 1.183 | 0.3682 | No | ||
| 62 | PXN | 3619 | 1.118 | 0.3626 | No | ||
| 63 | GNA13 | 3631 | 1.115 | 0.3667 | No | ||
| 64 | ARAF | 3649 | 1.111 | 0.3704 | No | ||
| 65 | ARHGEF7 | 3785 | 1.066 | 0.3676 | No | ||
| 66 | MRAS | 3933 | 1.021 | 0.3639 | No | ||
| 67 | RDX | 4010 | 1.001 | 0.3640 | No | ||
| 68 | APC2 | 4052 | 0.989 | 0.3660 | No | ||
| 69 | ITGAX | 4083 | 0.981 | 0.3685 | No | ||
| 70 | TMSB4X | 4183 | 0.954 | 0.3671 | No | ||
| 71 | NCKAP1 | 4617 | 0.842 | 0.3472 | No | ||
| 72 | ITGA2B | 4791 | 0.798 | 0.3411 | No | ||
| 73 | TIAM1 | 4866 | 0.778 | 0.3404 | No | ||
| 74 | PAK1 | 4985 | 0.751 | 0.3372 | No | ||
| 75 | RAC3 | 4987 | 0.751 | 0.3403 | No | ||
| 76 | MYL2 | 5017 | 0.744 | 0.3418 | No | ||
| 77 | CHRM4 | 5018 | 0.744 | 0.3450 | No | ||
| 78 | ARHGEF12 | 5034 | 0.741 | 0.3473 | No | ||
| 79 | TIAM2 | 5330 | 0.667 | 0.3341 | No | ||
| 80 | PFN3 | 5417 | 0.648 | 0.3321 | No | ||
| 81 | PDGFA | 5453 | 0.640 | 0.3329 | No | ||
| 82 | FGD3 | 5508 | 0.629 | 0.3327 | No | ||
| 83 | SOS1 | 5763 | 0.579 | 0.3213 | No | ||
| 84 | PIK3CD | 5865 | 0.555 | 0.3182 | No | ||
| 85 | MOS | 5993 | 0.526 | 0.3135 | No | ||
| 86 | FGF4 | 6104 | 0.500 | 0.3097 | No | ||
| 87 | PIK3R5 | 6156 | 0.490 | 0.3090 | No | ||
| 88 | CYFIP1 | 6322 | 0.454 | 0.3019 | No | ||
| 89 | ITGB3 | 6344 | 0.449 | 0.3027 | No | ||
| 90 | RRAS2 | 6520 | 0.408 | 0.2949 | No | ||
| 91 | ITGA6 | 6543 | 0.403 | 0.2954 | No | ||
| 92 | FGF20 | 7125 | 0.277 | 0.2651 | No | ||
| 93 | WASF1 | 7142 | 0.273 | 0.2653 | No | ||
| 94 | ITGAV | 7269 | 0.248 | 0.2595 | No | ||
| 95 | ITGA5 | 7284 | 0.245 | 0.2598 | No | ||
| 96 | CD14 | 7370 | 0.227 | 0.2562 | No | ||
| 97 | PIK3R1 | 7425 | 0.214 | 0.2541 | No | ||
| 98 | FGF2 | 7736 | 0.160 | 0.2380 | No | ||
| 99 | MYLK | 7818 | 0.144 | 0.2342 | No | ||
| 100 | PIK3R3 | 7845 | 0.138 | 0.2334 | No | ||
| 101 | GSN | 7935 | 0.119 | 0.2291 | No | ||
| 102 | FGF11 | 8002 | 0.108 | 0.2259 | No | ||
| 103 | PFN1 | 8160 | 0.080 | 0.2178 | No | ||
| 104 | FGF7 | 8345 | 0.045 | 0.2080 | No | ||
| 105 | MYL9 | 8368 | 0.041 | 0.2069 | No | ||
| 106 | LIMK1 | 8436 | 0.023 | 0.2034 | No | ||
| 107 | MYLC2PL | 8505 | 0.008 | 0.1998 | No | ||
| 108 | MYH10 | 8664 | -0.025 | 0.1913 | No | ||
| 109 | CHRM2 | 8858 | -0.061 | 0.1811 | No | ||
| 110 | ITGA4 | 8938 | -0.078 | 0.1771 | No | ||
| 111 | FGD1 | 8965 | -0.084 | 0.1761 | No | ||
| 112 | CHRM5 | 9140 | -0.123 | 0.1672 | No | ||
| 113 | FGF16 | 9182 | -0.130 | 0.1655 | No | ||
| 114 | ARPC4 | 9321 | -0.164 | 0.1587 | No | ||
| 115 | CHRM3 | 9370 | -0.171 | 0.1568 | No | ||
| 116 | PIK3R2 | 9527 | -0.203 | 0.1492 | No | ||
| 117 | NRAS | 9654 | -0.233 | 0.1434 | No | ||
| 118 | PAK7 | 10327 | -0.381 | 0.1085 | No | ||
| 119 | KRAS | 10637 | -0.440 | 0.0936 | No | ||
| 120 | MYLPF | 10772 | -0.469 | 0.0883 | No | ||
| 121 | ITGAM | 11004 | -0.520 | 0.0780 | No | ||
| 122 | ARPC5 | 11018 | -0.522 | 0.0795 | No | ||
| 123 | PDGFRA | 11164 | -0.546 | 0.0739 | No | ||
| 124 | GNA12 | 11199 | -0.554 | 0.0744 | No | ||
| 125 | ABI2 | 11227 | -0.561 | 0.0753 | No | ||
| 126 | EGFR | 11316 | -0.578 | 0.0729 | No | ||
| 127 | CFL1 | 11393 | -0.592 | 0.0713 | No | ||
| 128 | FGF23 | 11688 | -0.648 | 0.0581 | No | ||
| 129 | MYH14 | 11809 | -0.672 | 0.0544 | No | ||
| 130 | FGF21 | 12001 | -0.706 | 0.0470 | No | ||
| 131 | RAC1 | 12108 | -0.727 | 0.0444 | No | ||
| 132 | CRK | 12136 | -0.732 | 0.0460 | No | ||
| 133 | PPP1CB | 12344 | -0.780 | 0.0380 | No | ||
| 134 | PIP5K1B | 12408 | -0.792 | 0.0379 | No | ||
| 135 | PAK3 | 12559 | -0.827 | 0.0333 | No | ||
| 136 | ACTN2 | 12960 | -0.911 | 0.0154 | No | ||
| 137 | SCIN | 13135 | -0.949 | 0.0100 | No | ||
| 138 | HRAS | 13137 | -0.949 | 0.0139 | No | ||
| 139 | ITGA3 | 13324 | -0.993 | 0.0080 | No | ||
| 140 | FGF1 | 13522 | -1.034 | 0.0017 | No | ||
| 141 | ACTN3 | 13526 | -1.035 | 0.0059 | No | ||
| 142 | ITGB6 | 13576 | -1.048 | 0.0076 | No | ||
| 143 | ARPC3 | 13673 | -1.070 | 0.0069 | No | ||
| 144 | FGF12 | 13816 | -1.104 | 0.0039 | No | ||
| 145 | PIK3CB | 14076 | -1.164 | -0.0053 | No | ||
| 146 | PIK3CA | 14098 | -1.169 | -0.0015 | No | ||
| 147 | FGF18 | 14104 | -1.171 | 0.0032 | No | ||
| 148 | PPP1CA | 14222 | -1.201 | 0.0019 | No | ||
| 149 | DIAPH3 | 14333 | -1.228 | 0.0011 | No | ||
| 150 | ITGA7 | 14662 | -1.305 | -0.0112 | No | ||
| 151 | CRKL | 14828 | -1.344 | -0.0145 | No | ||
| 152 | CHRM1 | 14998 | -1.388 | -0.0178 | No | ||
| 153 | ITGA11 | 15122 | -1.417 | -0.0185 | No | ||
| 154 | SSH2 | 15380 | -1.495 | -0.0262 | No | ||
| 155 | FGF13 | 15436 | -1.513 | -0.0228 | No | ||
| 156 | PDGFB | 15771 | -1.618 | -0.0341 | No | ||
| 157 | PAK2 | 15809 | -1.628 | -0.0292 | No | ||
| 158 | FGF9 | 15892 | -1.659 | -0.0267 | No | ||
| 159 | BAIAP2 | 15925 | -1.671 | -0.0214 | No | ||
| 160 | ITGB4 | 16086 | -1.729 | -0.0228 | No | ||
| 161 | FGFR4 | 16240 | -1.776 | -0.0236 | No | ||
| 162 | FGF22 | 16420 | -1.838 | -0.0256 | No | ||
| 163 | PIP5K1A | 16432 | -1.845 | -0.0184 | No | ||
| 164 | MAPK1 | 16496 | -1.873 | -0.0139 | No | ||
| 165 | VAV3 | 16651 | -1.941 | -0.0141 | No | ||
| 166 | RHOA | 16825 | -2.021 | -0.0150 | No | ||
| 167 | F2 | 16859 | -2.037 | -0.0082 | No | ||
| 168 | ITGA8 | 17218 | -2.239 | -0.0182 | No | ||
| 169 | BCAR1 | 17300 | -2.290 | -0.0130 | No | ||
| 170 | PPP1CC | 17342 | -2.324 | -0.0054 | No | ||
| 171 | ITGAE | 17506 | -2.428 | -0.0040 | No | ||
| 172 | ITGB5 | 17533 | -2.442 | 0.0048 | No | ||
| 173 | PFN4 | 17640 | -2.546 | 0.0098 | No | ||
| 174 | F2R | 17972 | -2.869 | 0.0039 | No | ||
| 175 | ARPC1A | 18166 | -3.131 | 0.0067 | No | ||
| 176 | ITGA9 | 18512 | -4.204 | 0.0056 | No |
