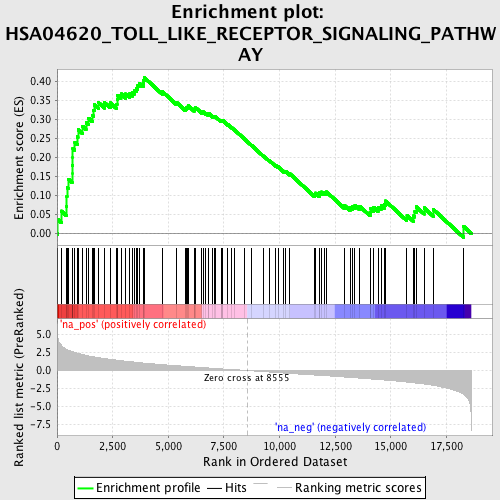
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HSA04620_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.4095662 |
| Normalized Enrichment Score (NES) | 1.8904604 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.04240028 |
| FWER p-Value | 0.328 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TIRAP | 35 | 4.471 | 0.0371 | Yes | ||
| 2 | MAP3K7IP2 | 188 | 3.487 | 0.0593 | Yes | ||
| 3 | TRAF3 | 428 | 2.923 | 0.0718 | Yes | ||
| 4 | MAPK14 | 438 | 2.913 | 0.0967 | Yes | ||
| 5 | IRAK1 | 447 | 2.898 | 0.1216 | Yes | ||
| 6 | MAP2K1 | 502 | 2.825 | 0.1433 | Yes | ||
| 7 | TBK1 | 676 | 2.610 | 0.1567 | Yes | ||
| 8 | IRF3 | 679 | 2.608 | 0.1793 | Yes | ||
| 9 | NFKB2 | 704 | 2.580 | 0.2005 | Yes | ||
| 10 | MAPK11 | 708 | 2.577 | 0.2228 | Yes | ||
| 11 | IKBKB | 798 | 2.484 | 0.2397 | Yes | ||
| 12 | IKBKG | 910 | 2.382 | 0.2544 | Yes | ||
| 13 | CD86 | 940 | 2.360 | 0.2734 | Yes | ||
| 14 | PIK3CG | 1139 | 2.200 | 0.2819 | Yes | ||
| 15 | MAPK3 | 1303 | 2.079 | 0.2912 | Yes | ||
| 16 | MAP3K7 | 1409 | 2.016 | 0.3031 | Yes | ||
| 17 | MAP2K6 | 1583 | 1.919 | 0.3105 | Yes | ||
| 18 | MAP2K2 | 1638 | 1.892 | 0.3241 | Yes | ||
| 19 | IFNA4 | 1676 | 1.868 | 0.3384 | Yes | ||
| 20 | IRF5 | 1866 | 1.776 | 0.3437 | Yes | ||
| 21 | TLR7 | 2131 | 1.658 | 0.3439 | Yes | ||
| 22 | MAPK9 | 2391 | 1.543 | 0.3433 | Yes | ||
| 23 | MAP3K7IP1 | 2681 | 1.433 | 0.3402 | Yes | ||
| 24 | TLR4 | 2695 | 1.430 | 0.3520 | Yes | ||
| 25 | MAP3K8 | 2710 | 1.426 | 0.3636 | Yes | ||
| 26 | IL6 | 2872 | 1.363 | 0.3668 | Yes | ||
| 27 | IFNA14 | 3085 | 1.296 | 0.3667 | Yes | ||
| 28 | TLR9 | 3270 | 1.230 | 0.3675 | Yes | ||
| 29 | IL12A | 3393 | 1.195 | 0.3713 | Yes | ||
| 30 | CHUK | 3488 | 1.162 | 0.3764 | Yes | ||
| 31 | TLR8 | 3571 | 1.136 | 0.3818 | Yes | ||
| 32 | RELA | 3628 | 1.116 | 0.3885 | Yes | ||
| 33 | NFKBIA | 3692 | 1.094 | 0.3947 | Yes | ||
| 34 | LY96 | 3869 | 1.037 | 0.3942 | Yes | ||
| 35 | MAP2K3 | 3890 | 1.029 | 0.4021 | Yes | ||
| 36 | CASP8 | 3918 | 1.024 | 0.4096 | Yes | ||
| 37 | IL1B | 4733 | 0.812 | 0.3727 | No | ||
| 38 | IFNAR2 | 5367 | 0.658 | 0.3443 | No | ||
| 39 | MAPK12 | 5765 | 0.579 | 0.3279 | No | ||
| 40 | IFNAR1 | 5810 | 0.569 | 0.3305 | No | ||
| 41 | PIK3CD | 5865 | 0.555 | 0.3324 | No | ||
| 42 | IRAK4 | 5892 | 0.551 | 0.3358 | No | ||
| 43 | PIK3R5 | 6156 | 0.490 | 0.3258 | No | ||
| 44 | TLR1 | 6193 | 0.484 | 0.3281 | No | ||
| 45 | CCL5 | 6229 | 0.475 | 0.3304 | No | ||
| 46 | TNF | 6499 | 0.413 | 0.3194 | No | ||
| 47 | SPP1 | 6562 | 0.399 | 0.3196 | No | ||
| 48 | IL12B | 6680 | 0.373 | 0.3165 | No | ||
| 49 | IFNA7 | 6805 | 0.347 | 0.3128 | No | ||
| 50 | RIPK1 | 6825 | 0.342 | 0.3148 | No | ||
| 51 | CXCL11 | 6999 | 0.303 | 0.3081 | No | ||
| 52 | MAPK13 | 7059 | 0.289 | 0.3074 | No | ||
| 53 | FOS | 7117 | 0.279 | 0.3068 | No | ||
| 54 | IRF7 | 7369 | 0.227 | 0.2952 | No | ||
| 55 | CD14 | 7370 | 0.227 | 0.2972 | No | ||
| 56 | PIK3R1 | 7425 | 0.214 | 0.2962 | No | ||
| 57 | LBP | 7452 | 0.208 | 0.2966 | No | ||
| 58 | CCL3 | 7675 | 0.171 | 0.2861 | No | ||
| 59 | PIK3R3 | 7845 | 0.138 | 0.2781 | No | ||
| 60 | IFNB1 | 7971 | 0.113 | 0.2724 | No | ||
| 61 | NFKB1 | 8403 | 0.033 | 0.2494 | No | ||
| 62 | TLR5 | 8754 | -0.041 | 0.2309 | No | ||
| 63 | AKT1 | 9294 | -0.158 | 0.2031 | No | ||
| 64 | PIK3R2 | 9527 | -0.203 | 0.1924 | No | ||
| 65 | CCL4 | 9816 | -0.269 | 0.1792 | No | ||
| 66 | IFNA13 | 9935 | -0.292 | 0.1754 | No | ||
| 67 | IKBKE | 10197 | -0.350 | 0.1643 | No | ||
| 68 | TICAM2 | 10258 | -0.365 | 0.1643 | No | ||
| 69 | TLR6 | 10458 | -0.406 | 0.1571 | No | ||
| 70 | CD80 | 11565 | -0.625 | 0.1028 | No | ||
| 71 | IFNA6 | 11598 | -0.632 | 0.1066 | No | ||
| 72 | STAT1 | 11783 | -0.667 | 0.1024 | No | ||
| 73 | CD40 | 11806 | -0.671 | 0.1071 | No | ||
| 74 | MAP2K7 | 11878 | -0.685 | 0.1093 | No | ||
| 75 | JUN | 12005 | -0.707 | 0.1086 | No | ||
| 76 | RAC1 | 12108 | -0.727 | 0.1094 | No | ||
| 77 | CXCL9 | 12926 | -0.904 | 0.0732 | No | ||
| 78 | MYD88 | 13171 | -0.957 | 0.0684 | No | ||
| 79 | IFNA1 | 13284 | -0.982 | 0.0709 | No | ||
| 80 | TOLLIP | 13385 | -1.005 | 0.0743 | No | ||
| 81 | TLR2 | 13599 | -1.055 | 0.0720 | No | ||
| 82 | PIK3CB | 14076 | -1.164 | 0.0564 | No | ||
| 83 | PIK3CA | 14098 | -1.169 | 0.0655 | No | ||
| 84 | MAP2K4 | 14231 | -1.203 | 0.0688 | No | ||
| 85 | MAPK10 | 14428 | -1.253 | 0.0692 | No | ||
| 86 | MAPK8 | 14563 | -1.284 | 0.0731 | No | ||
| 87 | FADD | 14702 | -1.315 | 0.0771 | No | ||
| 88 | AKT2 | 14759 | -1.328 | 0.0857 | No | ||
| 89 | CXCL10 | 15726 | -1.603 | 0.0475 | No | ||
| 90 | TLR3 | 16023 | -1.705 | 0.0464 | No | ||
| 91 | IFNA5 | 16081 | -1.727 | 0.0584 | No | ||
| 92 | IFNA2 | 16142 | -1.746 | 0.0703 | No | ||
| 93 | MAPK1 | 16496 | -1.873 | 0.0676 | No | ||
| 94 | TRAF6 | 16909 | -2.061 | 0.0633 | No | ||
| 95 | AKT3 | 18272 | -3.301 | 0.0186 | No |
