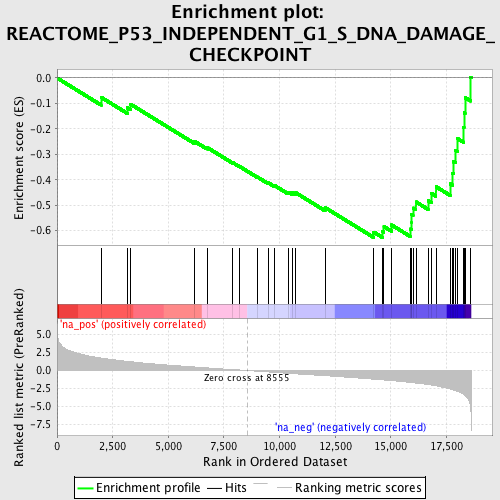
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | REACTOME_P53_INDEPENDENT_G1_S_DNA_DAMAGE_CHECKPOINT |
| Enrichment Score (ES) | -0.6268091 |
| Normalized Enrichment Score (NES) | -2.2284942 |
| Nominal p-value | 0.0 |
| FDR q-value | 2.499428E-4 |
| FWER p-Value | 0.0050 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PSMD11 | 1978 | 1.719 | -0.0760 | No | ||
| 2 | CDC25A | 3159 | 1.269 | -0.1170 | No | ||
| 3 | PSMC4 | 3311 | 1.219 | -0.1035 | No | ||
| 4 | PSMD3 | 6168 | 0.488 | -0.2485 | No | ||
| 5 | PSMD4 | 6740 | 0.360 | -0.2729 | No | ||
| 6 | PSMD7 | 7897 | 0.128 | -0.3328 | No | ||
| 7 | PSMA4 | 8185 | 0.076 | -0.3469 | No | ||
| 8 | PSMD8 | 8998 | -0.089 | -0.3890 | No | ||
| 9 | PSMB9 | 9493 | -0.199 | -0.4121 | No | ||
| 10 | PSMA5 | 9751 | -0.255 | -0.4214 | No | ||
| 11 | PSME1 | 10392 | -0.393 | -0.4489 | No | ||
| 12 | PSMD12 | 10560 | -0.426 | -0.4504 | No | ||
| 13 | PSME3 | 10728 | -0.459 | -0.4512 | No | ||
| 14 | PSMB10 | 12046 | -0.713 | -0.5094 | No | ||
| 15 | PSMC5 | 14228 | -1.202 | -0.6055 | Yes | ||
| 16 | PSMD14 | 14616 | -1.293 | -0.6034 | Yes | ||
| 17 | PSMC3 | 14692 | -1.312 | -0.5842 | Yes | ||
| 18 | PSMC2 | 15029 | -1.396 | -0.5775 | Yes | ||
| 19 | PSMB2 | 15870 | -1.651 | -0.5935 | Yes | ||
| 20 | PSMC6 | 15920 | -1.669 | -0.5665 | Yes | ||
| 21 | PSMD5 | 15926 | -1.671 | -0.5372 | Yes | ||
| 22 | PSMA6 | 15999 | -1.696 | -0.5110 | Yes | ||
| 23 | PSMC1 | 16131 | -1.742 | -0.4872 | Yes | ||
| 24 | PSMA1 | 16678 | -1.954 | -0.4819 | Yes | ||
| 25 | PSMA3 | 16833 | -2.024 | -0.4543 | Yes | ||
| 26 | PSMA2 | 17030 | -2.122 | -0.4273 | Yes | ||
| 27 | PSMB5 | 17668 | -2.567 | -0.4160 | Yes | ||
| 28 | PSMB1 | 17769 | -2.650 | -0.3745 | Yes | ||
| 29 | PSMA7 | 17818 | -2.692 | -0.3293 | Yes | ||
| 30 | PSMB4 | 17903 | -2.770 | -0.2847 | Yes | ||
| 31 | PSMD10 | 18007 | -2.903 | -0.2388 | Yes | ||
| 32 | PSMD2 | 18267 | -3.295 | -0.1944 | Yes | ||
| 33 | PSMB6 | 18293 | -3.344 | -0.1364 | Yes | ||
| 34 | PSMB3 | 18339 | -3.474 | -0.0773 | Yes | ||
| 35 | PSMD9 | 18587 | -5.197 | 0.0016 | Yes |