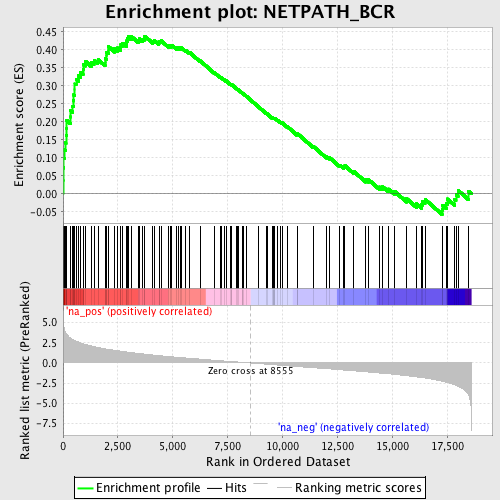
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_DMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | NETPATH_BCR |
| Enrichment Score (ES) | 0.4381206 |
| Normalized Enrichment Score (NES) | 2.0804203 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.016949704 |
| FWER p-Value | 0.059 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CARD11 | 1 | 6.382 | 0.0379 | Yes | ||
| 2 | CD22 | 4 | 6.009 | 0.0736 | Yes | ||
| 3 | BCL2 | 39 | 4.461 | 0.0983 | Yes | ||
| 4 | RAPGEF1 | 50 | 4.258 | 0.1231 | Yes | ||
| 5 | SYK | 115 | 3.756 | 0.1420 | Yes | ||
| 6 | DAPP1 | 133 | 3.682 | 0.1631 | Yes | ||
| 7 | WAS | 159 | 3.582 | 0.1830 | Yes | ||
| 8 | BTK | 166 | 3.564 | 0.2039 | Yes | ||
| 9 | DOK3 | 327 | 3.093 | 0.2137 | Yes | ||
| 10 | PTPN6 | 332 | 3.080 | 0.2318 | Yes | ||
| 11 | MAPK14 | 438 | 2.913 | 0.2435 | Yes | ||
| 12 | NFATC1 | 467 | 2.872 | 0.2591 | Yes | ||
| 13 | NFATC3 | 488 | 2.843 | 0.2749 | Yes | ||
| 14 | PIP5K1C | 539 | 2.776 | 0.2887 | Yes | ||
| 15 | LAT2 | 540 | 2.771 | 0.3052 | Yes | ||
| 16 | FCGR2B | 593 | 2.703 | 0.3185 | Yes | ||
| 17 | RPS6KA1 | 701 | 2.582 | 0.3281 | Yes | ||
| 18 | IKBKB | 798 | 2.484 | 0.3377 | Yes | ||
| 19 | IKBKG | 910 | 2.382 | 0.3459 | Yes | ||
| 20 | GRB2 | 924 | 2.371 | 0.3593 | Yes | ||
| 21 | RPS6KB1 | 1004 | 2.317 | 0.3688 | Yes | ||
| 22 | MAPK3 | 1303 | 2.079 | 0.3651 | Yes | ||
| 23 | MAP3K7 | 1409 | 2.016 | 0.3714 | Yes | ||
| 24 | CD19 | 1590 | 1.916 | 0.3731 | Yes | ||
| 25 | ATF2 | 1915 | 1.751 | 0.3660 | Yes | ||
| 26 | PRKCD | 1917 | 1.749 | 0.3764 | Yes | ||
| 27 | CSK | 1969 | 1.722 | 0.3839 | Yes | ||
| 28 | BLNK | 1972 | 1.721 | 0.3940 | Yes | ||
| 29 | CASP9 | 2060 | 1.688 | 0.3994 | Yes | ||
| 30 | VAV1 | 2063 | 1.688 | 0.4093 | Yes | ||
| 31 | CREB1 | 2352 | 1.562 | 0.4031 | Yes | ||
| 32 | PTK2 | 2477 | 1.514 | 0.4054 | Yes | ||
| 33 | SOS2 | 2609 | 1.463 | 0.4070 | Yes | ||
| 34 | CCNE1 | 2634 | 1.450 | 0.4143 | Yes | ||
| 35 | CDK2 | 2711 | 1.425 | 0.4187 | Yes | ||
| 36 | TEC | 2878 | 1.359 | 0.4178 | Yes | ||
| 37 | CCND2 | 2900 | 1.351 | 0.4247 | Yes | ||
| 38 | PPP3CA | 2926 | 1.342 | 0.4314 | Yes | ||
| 39 | VAV2 | 2980 | 1.326 | 0.4364 | Yes | ||
| 40 | CR2 | 3114 | 1.287 | 0.4369 | Yes | ||
| 41 | PTPRC | 3435 | 1.179 | 0.4266 | Yes | ||
| 42 | CHUK | 3488 | 1.162 | 0.4307 | Yes | ||
| 43 | RELA | 3628 | 1.116 | 0.4298 | Yes | ||
| 44 | NFKBIA | 3692 | 1.094 | 0.4330 | Yes | ||
| 45 | PLCG1 | 3717 | 1.086 | 0.4381 | Yes | ||
| 46 | CBL | 4089 | 0.980 | 0.4239 | No | ||
| 47 | NFATC2 | 4161 | 0.959 | 0.4258 | No | ||
| 48 | RASA1 | 4370 | 0.907 | 0.4199 | No | ||
| 49 | PDPK1 | 4391 | 0.901 | 0.4242 | No | ||
| 50 | PTK2B | 4470 | 0.877 | 0.4252 | No | ||
| 51 | NCK1 | 4808 | 0.793 | 0.4117 | No | ||
| 52 | CD5 | 4891 | 0.773 | 0.4119 | No | ||
| 53 | NEDD9 | 4950 | 0.759 | 0.4133 | No | ||
| 54 | CCND3 | 5169 | 0.709 | 0.4057 | No | ||
| 55 | CASP7 | 5238 | 0.693 | 0.4062 | No | ||
| 56 | HCLS1 | 5328 | 0.668 | 0.4053 | No | ||
| 57 | SH3BP2 | 5396 | 0.651 | 0.4056 | No | ||
| 58 | HCK | 5588 | 0.613 | 0.3989 | No | ||
| 59 | SOS1 | 5763 | 0.579 | 0.3930 | No | ||
| 60 | CD79A | 6252 | 0.470 | 0.3694 | No | ||
| 61 | ACTR3 | 6887 | 0.328 | 0.3371 | No | ||
| 62 | CDK6 | 7151 | 0.272 | 0.3245 | No | ||
| 63 | CBLB | 7208 | 0.260 | 0.3230 | No | ||
| 64 | PTPN18 | 7351 | 0.232 | 0.3167 | No | ||
| 65 | PIK3R1 | 7425 | 0.214 | 0.3140 | No | ||
| 66 | REL | 7630 | 0.179 | 0.3041 | No | ||
| 67 | RPS6 | 7655 | 0.174 | 0.3038 | No | ||
| 68 | ITPR2 | 7687 | 0.169 | 0.3032 | No | ||
| 69 | ITK | 7885 | 0.130 | 0.2933 | No | ||
| 70 | MAP4K1 | 7938 | 0.119 | 0.2912 | No | ||
| 71 | LYN | 7998 | 0.109 | 0.2886 | No | ||
| 72 | RB1 | 8189 | 0.075 | 0.2788 | No | ||
| 73 | PIK3AP1 | 8190 | 0.075 | 0.2793 | No | ||
| 74 | RASGRP3 | 8241 | 0.067 | 0.2770 | No | ||
| 75 | GSK3A | 8352 | 0.044 | 0.2713 | No | ||
| 76 | BCL6 | 8894 | -0.069 | 0.2424 | No | ||
| 77 | CCNA2 | 9253 | -0.149 | 0.2240 | No | ||
| 78 | AKT1 | 9294 | -0.158 | 0.2228 | No | ||
| 79 | PIK3R2 | 9527 | -0.203 | 0.2114 | No | ||
| 80 | CDK7 | 9570 | -0.214 | 0.2104 | No | ||
| 81 | CD72 | 9594 | -0.219 | 0.2105 | No | ||
| 82 | PRKCE | 9636 | -0.228 | 0.2096 | No | ||
| 83 | GAB2 | 9649 | -0.231 | 0.2104 | No | ||
| 84 | PRKCQ | 9793 | -0.264 | 0.2042 | No | ||
| 85 | GSK3B | 9910 | -0.287 | 0.1997 | No | ||
| 86 | CYCS | 9987 | -0.305 | 0.1974 | No | ||
| 87 | GAB1 | 10210 | -0.353 | 0.1875 | No | ||
| 88 | ACTR2 | 10660 | -0.444 | 0.1658 | No | ||
| 89 | MAPKAPK2 | 10672 | -0.448 | 0.1679 | No | ||
| 90 | CD79B | 11408 | -0.595 | 0.1317 | No | ||
| 91 | JUN | 12005 | -0.707 | 0.1037 | No | ||
| 92 | CRK | 12136 | -0.732 | 0.1011 | No | ||
| 93 | PRKD1 | 12618 | -0.841 | 0.0801 | No | ||
| 94 | HDAC5 | 12785 | -0.875 | 0.0763 | No | ||
| 95 | BCL2L11 | 12844 | -0.888 | 0.0785 | No | ||
| 96 | MAPK4 | 13237 | -0.973 | 0.0631 | No | ||
| 97 | PDK2 | 13795 | -1.099 | 0.0395 | No | ||
| 98 | FYN | 13920 | -1.129 | 0.0395 | No | ||
| 99 | ZAP70 | 14433 | -1.254 | 0.0193 | No | ||
| 100 | MAPK8 | 14563 | -1.284 | 0.0200 | No | ||
| 101 | CRKL | 14828 | -1.344 | 0.0137 | No | ||
| 102 | SHC1 | 15101 | -1.412 | 0.0074 | No | ||
| 103 | PPP3R1 | 15658 | -1.582 | -0.0132 | No | ||
| 104 | BCL10 | 16112 | -1.737 | -0.0273 | No | ||
| 105 | DOK1 | 16346 | -1.816 | -0.0291 | No | ||
| 106 | BLK | 16383 | -1.826 | -0.0202 | No | ||
| 107 | MAPK1 | 16496 | -1.873 | -0.0151 | No | ||
| 108 | CD81 | 17273 | -2.273 | -0.0435 | No | ||
| 109 | BCAR1 | 17300 | -2.290 | -0.0313 | No | ||
| 110 | LCP2 | 17473 | -2.411 | -0.0262 | No | ||
| 111 | PTPN11 | 17514 | -2.432 | -0.0139 | No | ||
| 112 | LCK | 17843 | -2.721 | -0.0154 | No | ||
| 113 | GTF2I | 17907 | -2.775 | -0.0023 | No | ||
| 114 | BAX | 18002 | -2.899 | 0.0099 | No | ||
| 115 | CDK4 | 18466 | -3.907 | 0.0081 | No |
