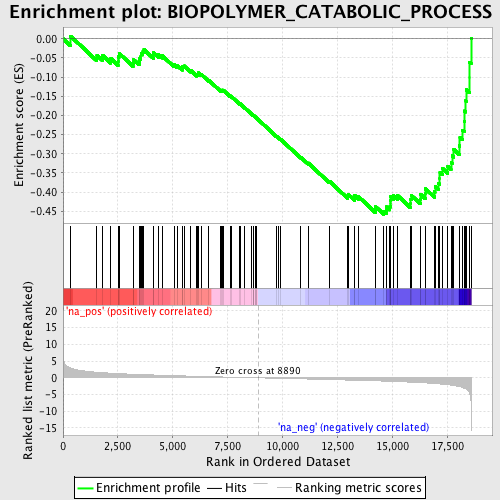
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | BIOPOLYMER_CATABOLIC_PROCESS |
| Enrichment Score (ES) | -0.45857465 |
| Normalized Enrichment Score (NES) | -1.8624899 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07118003 |
| FWER p-Value | 0.636 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MGAM | 342 | 2.884 | 0.0071 | No | ||
| 2 | SMG6 | 1541 | 1.654 | -0.0430 | No | ||
| 3 | NEDD8 | 1796 | 1.534 | -0.0431 | No | ||
| 4 | CHIT1 | 2175 | 1.385 | -0.0512 | No | ||
| 5 | ZFP36 | 2507 | 1.272 | -0.0578 | No | ||
| 6 | PYGB | 2514 | 1.269 | -0.0469 | No | ||
| 7 | EDEM1 | 2549 | 1.258 | -0.0376 | No | ||
| 8 | POP1 | 3190 | 1.074 | -0.0626 | No | ||
| 9 | ISG20 | 3221 | 1.065 | -0.0548 | No | ||
| 10 | UBE3A | 3471 | 0.998 | -0.0594 | No | ||
| 11 | EGLN2 | 3494 | 0.992 | -0.0518 | No | ||
| 12 | UBE2B | 3545 | 0.976 | -0.0459 | No | ||
| 13 | UBB | 3549 | 0.975 | -0.0374 | No | ||
| 14 | AMFR | 3639 | 0.953 | -0.0338 | No | ||
| 15 | MDM2 | 3684 | 0.943 | -0.0278 | No | ||
| 16 | UBE4A | 4120 | 0.847 | -0.0438 | No | ||
| 17 | CDC23 | 4122 | 0.846 | -0.0363 | No | ||
| 18 | UPF2 | 4343 | 0.790 | -0.0412 | No | ||
| 19 | UBE2N | 4513 | 0.753 | -0.0437 | No | ||
| 20 | UBE2D2 | 5060 | 0.648 | -0.0674 | No | ||
| 21 | SYVN1 | 5218 | 0.617 | -0.0704 | No | ||
| 22 | RNASEH1 | 5426 | 0.579 | -0.0765 | No | ||
| 23 | UBE4B | 5435 | 0.577 | -0.0718 | No | ||
| 24 | DFFA | 5509 | 0.563 | -0.0708 | No | ||
| 25 | CECR2 | 5807 | 0.507 | -0.0823 | No | ||
| 26 | FZR1 | 6087 | 0.458 | -0.0933 | No | ||
| 27 | MDM4 | 6141 | 0.449 | -0.0922 | No | ||
| 28 | CDKN2A | 6152 | 0.447 | -0.0888 | No | ||
| 29 | UFD1L | 6299 | 0.418 | -0.0930 | No | ||
| 30 | FBXL4 | 6637 | 0.365 | -0.1079 | No | ||
| 31 | UBE2A | 7195 | 0.267 | -0.1356 | No | ||
| 32 | ERN2 | 7226 | 0.263 | -0.1349 | No | ||
| 33 | UBE2G2 | 7244 | 0.259 | -0.1335 | No | ||
| 34 | HNRPD | 7296 | 0.251 | -0.1341 | No | ||
| 35 | TSG101 | 7607 | 0.204 | -0.1490 | No | ||
| 36 | SMURF2 | 7674 | 0.193 | -0.1509 | No | ||
| 37 | PSMD14 | 8041 | 0.132 | -0.1695 | No | ||
| 38 | FBXO7 | 8065 | 0.127 | -0.1696 | No | ||
| 39 | SIAH2 | 8274 | 0.099 | -0.1799 | No | ||
| 40 | UBE2C | 8567 | 0.052 | -0.1952 | No | ||
| 41 | ARIH1 | 8662 | 0.038 | -0.2000 | No | ||
| 42 | FOXL2 | 8674 | 0.037 | -0.2002 | No | ||
| 43 | UBE2D3 | 8697 | 0.033 | -0.2011 | No | ||
| 44 | RNASE2 | 8745 | 0.026 | -0.2034 | No | ||
| 45 | USP33 | 8747 | 0.025 | -0.2033 | No | ||
| 46 | HGS | 8812 | 0.015 | -0.2066 | No | ||
| 47 | ANAPC4 | 8813 | 0.015 | -0.2064 | No | ||
| 48 | SOD1 | 9720 | -0.123 | -0.2543 | No | ||
| 49 | BTRC | 9809 | -0.136 | -0.2578 | No | ||
| 50 | TPD52L1 | 9925 | -0.155 | -0.2626 | No | ||
| 51 | SMURF1 | 10827 | -0.296 | -0.3087 | No | ||
| 52 | CIDEA | 11173 | -0.355 | -0.3242 | No | ||
| 53 | BNIP3 | 12140 | -0.511 | -0.3718 | No | ||
| 54 | GAA | 12980 | -0.664 | -0.4112 | No | ||
| 55 | UBE2I | 12997 | -0.667 | -0.4062 | No | ||
| 56 | ERCC3 | 13287 | -0.711 | -0.4155 | No | ||
| 57 | PSMC5 | 13290 | -0.711 | -0.4093 | No | ||
| 58 | ERCC4 | 13466 | -0.742 | -0.4122 | No | ||
| 59 | UBE2D1 | 14235 | -0.890 | -0.4457 | No | ||
| 60 | UBE2V2 | 14250 | -0.893 | -0.4386 | No | ||
| 61 | CYCS | 14621 | -0.973 | -0.4500 | Yes | ||
| 62 | PPP1R8 | 14728 | -0.996 | -0.4469 | Yes | ||
| 63 | XRN2 | 14741 | -0.998 | -0.4387 | Yes | ||
| 64 | RNF11 | 14894 | -1.035 | -0.4377 | Yes | ||
| 65 | DFFB | 14923 | -1.042 | -0.4300 | Yes | ||
| 66 | ANAPC10 | 14925 | -1.042 | -0.4208 | Yes | ||
| 67 | RNASE6 | 14934 | -1.044 | -0.4120 | Yes | ||
| 68 | PCNP | 15058 | -1.072 | -0.4091 | Yes | ||
| 69 | RNASEH2A | 15246 | -1.124 | -0.4093 | Yes | ||
| 70 | ABCE1 | 15839 | -1.279 | -0.4299 | Yes | ||
| 71 | UBE2G1 | 15846 | -1.282 | -0.4189 | Yes | ||
| 72 | ANAPC2 | 15868 | -1.288 | -0.4086 | Yes | ||
| 73 | AIFM1 | 16280 | -1.420 | -0.4182 | Yes | ||
| 74 | NTHL1 | 16291 | -1.425 | -0.4061 | Yes | ||
| 75 | GSPT1 | 16507 | -1.506 | -0.4044 | Yes | ||
| 76 | PRSS2 | 16516 | -1.509 | -0.3915 | Yes | ||
| 77 | FBXO22 | 16925 | -1.683 | -0.3986 | Yes | ||
| 78 | PABPC4 | 16951 | -1.695 | -0.3849 | Yes | ||
| 79 | AUH | 17113 | -1.788 | -0.3778 | Yes | ||
| 80 | STUB1 | 17170 | -1.813 | -0.3648 | Yes | ||
| 81 | PARK2 | 17175 | -1.816 | -0.3489 | Yes | ||
| 82 | FRAP1 | 17284 | -1.890 | -0.3380 | Yes | ||
| 83 | ERCC5 | 17532 | -2.048 | -0.3332 | Yes | ||
| 84 | UBE2H | 17683 | -2.162 | -0.3221 | Yes | ||
| 85 | ERCC2 | 17734 | -2.206 | -0.3053 | Yes | ||
| 86 | UBE2L3 | 17807 | -2.273 | -0.2890 | Yes | ||
| 87 | ERCC1 | 18056 | -2.606 | -0.2793 | Yes | ||
| 88 | SQSTM1 | 18087 | -2.668 | -0.2573 | Yes | ||
| 89 | CDC20 | 18204 | -2.877 | -0.2381 | Yes | ||
| 90 | FAF1 | 18286 | -3.081 | -0.2152 | Yes | ||
| 91 | UBE2E1 | 18298 | -3.108 | -0.1883 | Yes | ||
| 92 | VCP | 18353 | -3.305 | -0.1619 | Yes | ||
| 93 | ANAPC5 | 18372 | -3.380 | -0.1329 | Yes | ||
| 94 | UHRF2 | 18521 | -4.493 | -0.1011 | Yes | ||
| 95 | PPT1 | 18522 | -4.500 | -0.0613 | Yes | ||
| 96 | BAX | 18598 | -7.485 | 0.0010 | Yes |
