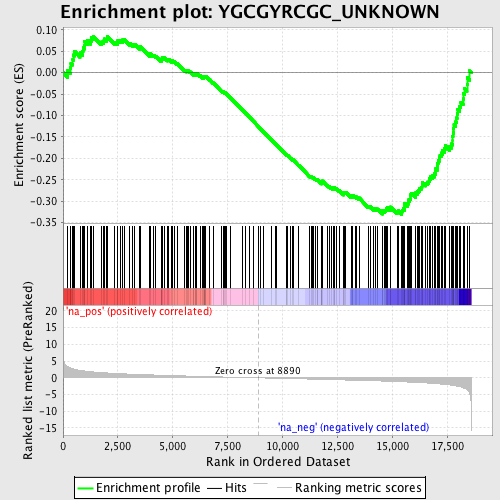
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | YGCGYRCGC_UNKNOWN |
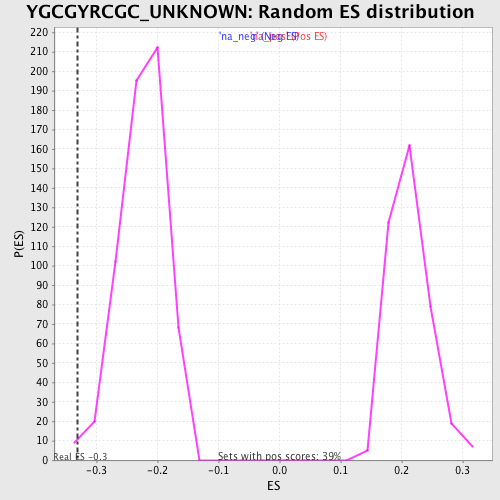
| Enrichment Score (ES) | -0.33096877 |
| Normalized Enrichment Score (NES) | -1.4801949 |
| Nominal p-value | 0.004950495 |
| FDR q-value | 0.4214905 |
| FWER p-Value | 0.996 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MAPT | 202 | 3.363 | 0.0043 | No | ||
| 2 | CDK5R1 | 320 | 2.949 | 0.0114 | No | ||
| 3 | POU2F1 | 356 | 2.857 | 0.0225 | No | ||
| 4 | CYP1B1 | 444 | 2.691 | 0.0300 | No | ||
| 5 | KLF13 | 459 | 2.658 | 0.0414 | No | ||
| 6 | ARRB2 | 508 | 2.577 | 0.0505 | No | ||
| 7 | RRAS | 770 | 2.216 | 0.0464 | No | ||
| 8 | CCS | 891 | 2.099 | 0.0494 | No | ||
| 9 | TMEM24 | 909 | 2.078 | 0.0580 | No | ||
| 10 | HNRPDL | 963 | 2.042 | 0.0644 | No | ||
| 11 | HOOK3 | 985 | 2.020 | 0.0724 | No | ||
| 12 | TRIM8 | 1104 | 1.926 | 0.0748 | No | ||
| 13 | HIC2 | 1256 | 1.831 | 0.0749 | No | ||
| 14 | ATP2A2 | 1283 | 1.815 | 0.0818 | No | ||
| 15 | WBSCR17 | 1366 | 1.756 | 0.0853 | No | ||
| 16 | DLGAP4 | 1748 | 1.552 | 0.0717 | No | ||
| 17 | TIMELESS | 1834 | 1.519 | 0.0740 | No | ||
| 18 | NAGK | 1873 | 1.503 | 0.0787 | No | ||
| 19 | UBTF | 1999 | 1.452 | 0.0786 | No | ||
| 20 | DDIT3 | 2006 | 1.449 | 0.0848 | No | ||
| 21 | GBX2 | 2363 | 1.316 | 0.0715 | No | ||
| 22 | SLC25A11 | 2474 | 1.283 | 0.0714 | No | ||
| 23 | MECP2 | 2497 | 1.275 | 0.0760 | No | ||
| 24 | JPH4 | 2599 | 1.242 | 0.0761 | No | ||
| 25 | TIAM1 | 2696 | 1.211 | 0.0764 | No | ||
| 26 | SLC16A6 | 2781 | 1.188 | 0.0773 | No | ||
| 27 | KCNC1 | 3028 | 1.115 | 0.0690 | No | ||
| 28 | MLLT10 | 3164 | 1.081 | 0.0666 | No | ||
| 29 | SLC35B3 | 3249 | 1.057 | 0.0668 | No | ||
| 30 | AKAP8 | 3474 | 0.997 | 0.0592 | No | ||
| 31 | CFL1 | 3509 | 0.989 | 0.0619 | No | ||
| 32 | VKORC1L1 | 3952 | 0.884 | 0.0419 | No | ||
| 33 | NXPH3 | 3972 | 0.881 | 0.0449 | No | ||
| 34 | CRSP7 | 4123 | 0.846 | 0.0406 | No | ||
| 35 | RAB10 | 4224 | 0.818 | 0.0389 | No | ||
| 36 | NPAS2 | 4470 | 0.763 | 0.0291 | No | ||
| 37 | MAP3K11 | 4497 | 0.756 | 0.0311 | No | ||
| 38 | KCNS2 | 4505 | 0.755 | 0.0341 | No | ||
| 39 | PMP22 | 4537 | 0.749 | 0.0359 | No | ||
| 40 | WAC | 4622 | 0.732 | 0.0346 | No | ||
| 41 | PCTK1 | 4761 | 0.705 | 0.0304 | No | ||
| 42 | SHH | 4804 | 0.695 | 0.0312 | No | ||
| 43 | PIM2 | 4930 | 0.673 | 0.0275 | No | ||
| 44 | GOPC | 4975 | 0.664 | 0.0281 | No | ||
| 45 | SLITRK4 | 5067 | 0.645 | 0.0261 | No | ||
| 46 | STARD10 | 5194 | 0.621 | 0.0221 | No | ||
| 47 | H1FX | 5535 | 0.557 | 0.0062 | No | ||
| 48 | GOLGA2 | 5623 | 0.543 | 0.0039 | No | ||
| 49 | KAZALD1 | 5660 | 0.536 | 0.0044 | No | ||
| 50 | MNT | 5709 | 0.527 | 0.0042 | No | ||
| 51 | UBE2S | 5798 | 0.509 | 0.0018 | No | ||
| 52 | NEK8 | 5941 | 0.484 | -0.0037 | No | ||
| 53 | BCL9 | 5956 | 0.482 | -0.0023 | No | ||
| 54 | POLR2B | 6047 | 0.465 | -0.0051 | No | ||
| 55 | RAB33A | 6053 | 0.464 | -0.0032 | No | ||
| 56 | DNAJA1 | 6059 | 0.463 | -0.0014 | No | ||
| 57 | ADAM12 | 6095 | 0.457 | -0.0012 | No | ||
| 58 | HNRPL | 6239 | 0.430 | -0.0070 | No | ||
| 59 | TPR | 6352 | 0.409 | -0.0113 | No | ||
| 60 | SORCS3 | 6416 | 0.398 | -0.0129 | No | ||
| 61 | CCNE1 | 6421 | 0.397 | -0.0113 | No | ||
| 62 | RPP38 | 6434 | 0.395 | -0.0101 | No | ||
| 63 | RBKS | 6451 | 0.391 | -0.0092 | No | ||
| 64 | NEURL | 6470 | 0.389 | -0.0084 | No | ||
| 65 | PACRG | 6509 | 0.382 | -0.0087 | No | ||
| 66 | SFRS6 | 6689 | 0.356 | -0.0168 | No | ||
| 67 | ATP5G2 | 6846 | 0.331 | -0.0238 | No | ||
| 68 | RQCD1 | 7217 | 0.263 | -0.0427 | No | ||
| 69 | LENG8 | 7288 | 0.252 | -0.0453 | No | ||
| 70 | HNRPD | 7296 | 0.251 | -0.0446 | No | ||
| 71 | KCNN1 | 7370 | 0.238 | -0.0475 | No | ||
| 72 | MST1 | 7381 | 0.237 | -0.0469 | No | ||
| 73 | ASCL1 | 7444 | 0.229 | -0.0492 | No | ||
| 74 | NR2F2 | 7629 | 0.200 | -0.0583 | No | ||
| 75 | FMR1 | 8166 | 0.114 | -0.0869 | No | ||
| 76 | PCBP2 | 8324 | 0.091 | -0.0950 | No | ||
| 77 | PRKAG2 | 8515 | 0.059 | -0.1050 | No | ||
| 78 | STX12 | 8699 | 0.033 | -0.1148 | No | ||
| 79 | PRKAR2A | 8899 | -0.001 | -0.1256 | No | ||
| 80 | POP7 | 8994 | -0.015 | -0.1307 | No | ||
| 81 | PAQR4 | 9149 | -0.039 | -0.1388 | No | ||
| 82 | TFRC | 9505 | -0.090 | -0.1577 | No | ||
| 83 | UROD | 9675 | -0.115 | -0.1663 | No | ||
| 84 | MMP16 | 9716 | -0.122 | -0.1680 | No | ||
| 85 | PPARGC1B | 10168 | -0.190 | -0.1916 | No | ||
| 86 | HNRPR | 10189 | -0.194 | -0.1918 | No | ||
| 87 | SLC12A5 | 10236 | -0.200 | -0.1934 | No | ||
| 88 | YWHAE | 10376 | -0.220 | -0.1999 | No | ||
| 89 | XPO5 | 10463 | -0.234 | -0.2035 | No | ||
| 90 | AATF | 10480 | -0.238 | -0.2033 | No | ||
| 91 | HOXA4 | 10744 | -0.283 | -0.2163 | No | ||
| 92 | CTDSPL | 11248 | -0.369 | -0.2419 | No | ||
| 93 | CRY1 | 11301 | -0.378 | -0.2430 | No | ||
| 94 | FBXW9 | 11353 | -0.385 | -0.2440 | No | ||
| 95 | RPS6KA5 | 11404 | -0.394 | -0.2449 | No | ||
| 96 | APRIN | 11500 | -0.407 | -0.2482 | No | ||
| 97 | NUDT3 | 11577 | -0.420 | -0.2505 | No | ||
| 98 | SEMA6A | 11585 | -0.421 | -0.2489 | No | ||
| 99 | A2BP1 | 11787 | -0.453 | -0.2578 | No | ||
| 100 | ZIC2 | 11788 | -0.453 | -0.2557 | No | ||
| 101 | LRFN5 | 11798 | -0.455 | -0.2541 | No | ||
| 102 | ACCN2 | 11810 | -0.457 | -0.2527 | No | ||
| 103 | CALM3 | 12042 | -0.493 | -0.2629 | No | ||
| 104 | NR2C2 | 12119 | -0.507 | -0.2648 | No | ||
| 105 | CKS2 | 12225 | -0.526 | -0.2681 | No | ||
| 106 | CDON | 12312 | -0.541 | -0.2703 | No | ||
| 107 | JMJD2C | 12360 | -0.550 | -0.2703 | No | ||
| 108 | GABRA4 | 12374 | -0.552 | -0.2685 | No | ||
| 109 | BRE | 12439 | -0.562 | -0.2694 | No | ||
| 110 | FUT8 | 12590 | -0.595 | -0.2749 | No | ||
| 111 | HNRPAB | 12788 | -0.626 | -0.2827 | No | ||
| 112 | FHOD1 | 12789 | -0.626 | -0.2799 | No | ||
| 113 | ROM1 | 12837 | -0.634 | -0.2795 | No | ||
| 114 | FUS | 12884 | -0.643 | -0.2791 | No | ||
| 115 | RAB6A | 13122 | -0.686 | -0.2889 | No | ||
| 116 | AOF2 | 13171 | -0.694 | -0.2883 | No | ||
| 117 | IMMT | 13181 | -0.696 | -0.2856 | No | ||
| 118 | MLLT7 | 13305 | -0.714 | -0.2891 | No | ||
| 119 | DLL1 | 13383 | -0.726 | -0.2899 | No | ||
| 120 | BCLAF1 | 13491 | -0.748 | -0.2923 | No | ||
| 121 | PGM2L1 | 13917 | -0.825 | -0.3117 | No | ||
| 122 | SYNCRIP | 13996 | -0.841 | -0.3121 | No | ||
| 123 | TRIB2 | 14161 | -0.874 | -0.3170 | No | ||
| 124 | PDE7A | 14249 | -0.892 | -0.3176 | No | ||
| 125 | SPRY4 | 14334 | -0.911 | -0.3181 | No | ||
| 126 | PPM1A | 14555 | -0.959 | -0.3256 | Yes | ||
| 127 | ZNHIT1 | 14565 | -0.960 | -0.3218 | Yes | ||
| 128 | GTF2A1 | 14644 | -0.978 | -0.3215 | Yes | ||
| 129 | RHOA | 14709 | -0.991 | -0.3205 | Yes | ||
| 130 | GEMIN4 | 14736 | -0.998 | -0.3174 | Yes | ||
| 131 | ZDHHC14 | 14774 | -1.008 | -0.3148 | Yes | ||
| 132 | U2AF2 | 14921 | -1.041 | -0.3180 | Yes | ||
| 133 | ANAPC10 | 14925 | -1.042 | -0.3134 | Yes | ||
| 134 | POU4F1 | 15224 | -1.118 | -0.3245 | Yes | ||
| 135 | TADA2L | 15280 | -1.131 | -0.3223 | Yes | ||
| 136 | PLOD3 | 15440 | -1.173 | -0.3256 | Yes | ||
| 137 | ACACA | 15446 | -1.174 | -0.3206 | Yes | ||
| 138 | SIX1 | 15491 | -1.187 | -0.3176 | Yes | ||
| 139 | ENDOGL1 | 15557 | -1.205 | -0.3156 | Yes | ||
| 140 | PSMD2 | 15560 | -1.205 | -0.3102 | Yes | ||
| 141 | COL25A1 | 15564 | -1.207 | -0.3049 | Yes | ||
| 142 | RBBP4 | 15708 | -1.241 | -0.3070 | Yes | ||
| 143 | NEUROD1 | 15723 | -1.246 | -0.3021 | Yes | ||
| 144 | MPP5 | 15735 | -1.249 | -0.2970 | Yes | ||
| 145 | MAZ | 15802 | -1.268 | -0.2948 | Yes | ||
| 146 | ABCE1 | 15839 | -1.279 | -0.2910 | Yes | ||
| 147 | RNF149 | 15841 | -1.280 | -0.2852 | Yes | ||
| 148 | ZZZ3 | 15885 | -1.292 | -0.2817 | Yes | ||
| 149 | MTMR3 | 16068 | -1.348 | -0.2854 | Yes | ||
| 150 | TIGD2 | 16078 | -1.352 | -0.2797 | Yes | ||
| 151 | SPINT2 | 16149 | -1.372 | -0.2773 | Yes | ||
| 152 | MTHFD2 | 16207 | -1.395 | -0.2740 | Yes | ||
| 153 | PHYHIPL | 16257 | -1.411 | -0.2703 | Yes | ||
| 154 | EN1 | 16316 | -1.435 | -0.2669 | Yes | ||
| 155 | IKBKAP | 16358 | -1.448 | -0.2625 | Yes | ||
| 156 | HOXA1 | 16362 | -1.450 | -0.2561 | Yes | ||
| 157 | CHD4 | 16518 | -1.510 | -0.2576 | Yes | ||
| 158 | MRPL14 | 16602 | -1.543 | -0.2551 | Yes | ||
| 159 | RUTBC3 | 16676 | -1.572 | -0.2519 | Yes | ||
| 160 | AP3S1 | 16711 | -1.588 | -0.2466 | Yes | ||
| 161 | SOX12 | 16760 | -1.607 | -0.2419 | Yes | ||
| 162 | TCTA | 16850 | -1.650 | -0.2392 | Yes | ||
| 163 | SLC25A12 | 16933 | -1.688 | -0.2360 | Yes | ||
| 164 | TRIM33 | 16978 | -1.708 | -0.2306 | Yes | ||
| 165 | RPS4X | 16989 | -1.717 | -0.2233 | Yes | ||
| 166 | TUBG1 | 17065 | -1.759 | -0.2194 | Yes | ||
| 167 | RGS7 | 17077 | -1.766 | -0.2119 | Yes | ||
| 168 | RNF38 | 17092 | -1.776 | -0.2046 | Yes | ||
| 169 | GTF2I | 17137 | -1.800 | -0.1988 | Yes | ||
| 170 | PARK2 | 17175 | -1.816 | -0.1926 | Yes | ||
| 171 | BAT3 | 17235 | -1.857 | -0.1873 | Yes | ||
| 172 | IMMP2L | 17296 | -1.896 | -0.1819 | Yes | ||
| 173 | TSGA10 | 17370 | -1.934 | -0.1771 | Yes | ||
| 174 | AAMP | 17411 | -1.958 | -0.1704 | Yes | ||
| 175 | EIF3S7 | 17617 | -2.120 | -0.1719 | Yes | ||
| 176 | ULK2 | 17696 | -2.173 | -0.1662 | Yes | ||
| 177 | USP37 | 17747 | -2.220 | -0.1588 | Yes | ||
| 178 | SEP15 | 17750 | -2.222 | -0.1488 | Yes | ||
| 179 | HS2ST1 | 17772 | -2.240 | -0.1398 | Yes | ||
| 180 | POLH | 17802 | -2.268 | -0.1310 | Yes | ||
| 181 | INHBB | 17814 | -2.278 | -0.1213 | Yes | ||
| 182 | DCTN4 | 17897 | -2.383 | -0.1149 | Yes | ||
| 183 | ACSL4 | 17930 | -2.422 | -0.1056 | Yes | ||
| 184 | HNRPH1 | 17956 | -2.455 | -0.0958 | Yes | ||
| 185 | SRR | 17990 | -2.510 | -0.0861 | Yes | ||
| 186 | VRK3 | 18080 | -2.651 | -0.0789 | Yes | ||
| 187 | SKP2 | 18129 | -2.730 | -0.0691 | Yes | ||
| 188 | PPP3CC | 18231 | -2.935 | -0.0612 | Yes | ||
| 189 | MRPL45 | 18242 | -2.955 | -0.0483 | Yes | ||
| 190 | ANP32A | 18303 | -3.120 | -0.0374 | Yes | ||
| 191 | HIBADH | 18409 | -3.532 | -0.0270 | Yes | ||
| 192 | MAGI1 | 18422 | -3.588 | -0.0113 | Yes | ||
| 193 | GNAS | 18538 | -4.797 | 0.0042 | Yes |