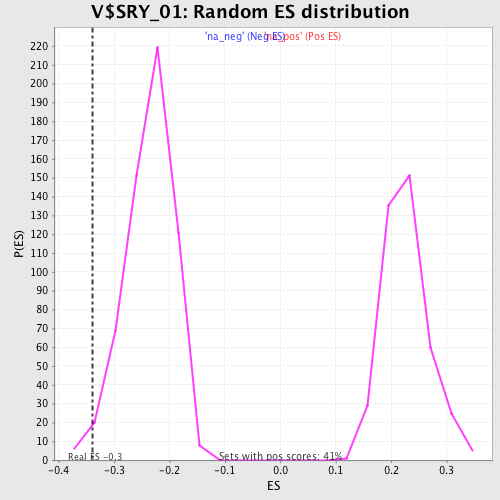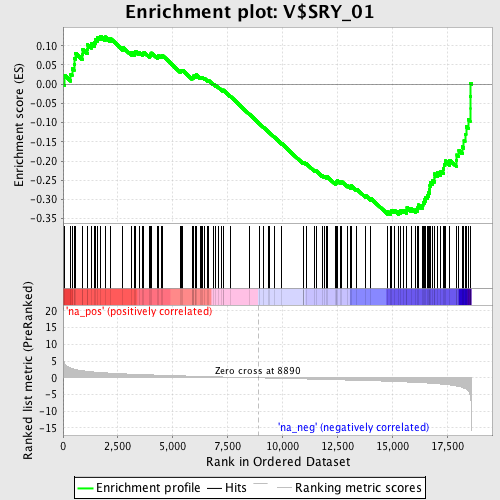
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$SRY_01 |
| Enrichment Score (ES) | -0.33910638 |
| Normalized Enrichment Score (NES) | -1.4373827 |
| Nominal p-value | 0.016835017 |
| FDR q-value | 0.5213024 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ATF7IP | 81 | 4.204 | 0.0224 | No | ||
| 2 | TCF7L2 | 348 | 2.868 | 0.0263 | No | ||
| 3 | CDKN1C | 410 | 2.747 | 0.0405 | No | ||
| 4 | ARID4B | 501 | 2.590 | 0.0522 | No | ||
| 5 | SORL1 | 518 | 2.560 | 0.0676 | No | ||
| 6 | FCHSD2 | 568 | 2.488 | 0.0808 | No | ||
| 7 | CTSK | 871 | 2.127 | 0.0781 | No | ||
| 8 | SP1 | 898 | 2.091 | 0.0900 | No | ||
| 9 | TRIM8 | 1104 | 1.926 | 0.0912 | No | ||
| 10 | PRRX1 | 1117 | 1.920 | 0.1028 | No | ||
| 11 | ATP2A2 | 1283 | 1.815 | 0.1054 | No | ||
| 12 | CYR61 | 1430 | 1.718 | 0.1085 | No | ||
| 13 | CACNA1E | 1471 | 1.693 | 0.1171 | No | ||
| 14 | GRIN2B | 1577 | 1.639 | 0.1219 | No | ||
| 15 | SOX5 | 1703 | 1.576 | 0.1252 | No | ||
| 16 | SORT1 | 1916 | 1.484 | 0.1232 | No | ||
| 17 | HLX1 | 2158 | 1.394 | 0.1190 | No | ||
| 18 | ATF7 | 2725 | 1.203 | 0.0961 | No | ||
| 19 | CRY2 | 3104 | 1.097 | 0.0826 | No | ||
| 20 | DAB1 | 3243 | 1.059 | 0.0819 | No | ||
| 21 | CYP26A1 | 3306 | 1.043 | 0.0852 | No | ||
| 22 | TAL1 | 3464 | 0.999 | 0.0831 | No | ||
| 23 | GFRA1 | 3637 | 0.955 | 0.0799 | No | ||
| 24 | HOXC6 | 3681 | 0.944 | 0.0836 | No | ||
| 25 | NFIA | 3929 | 0.889 | 0.0759 | No | ||
| 26 | PCTK2 | 3980 | 0.879 | 0.0788 | No | ||
| 27 | H3F3A | 4036 | 0.865 | 0.0813 | No | ||
| 28 | SESN2 | 4295 | 0.799 | 0.0725 | No | ||
| 29 | PVRL1 | 4339 | 0.791 | 0.0752 | No | ||
| 30 | SOX6 | 4461 | 0.765 | 0.0735 | No | ||
| 31 | SELENBP1 | 4520 | 0.752 | 0.0752 | No | ||
| 32 | JMJD1C | 5351 | 0.592 | 0.0341 | No | ||
| 33 | HTR3B | 5393 | 0.584 | 0.0356 | No | ||
| 34 | PCF11 | 5448 | 0.573 | 0.0363 | No | ||
| 35 | LDB1 | 5875 | 0.494 | 0.0164 | No | ||
| 36 | TEAD1 | 5892 | 0.491 | 0.0187 | No | ||
| 37 | LALBA | 5949 | 0.483 | 0.0187 | No | ||
| 38 | BCL9 | 5956 | 0.482 | 0.0215 | No | ||
| 39 | ENTPD1 | 6036 | 0.467 | 0.0202 | No | ||
| 40 | LRP5 | 6043 | 0.465 | 0.0228 | No | ||
| 41 | HMGN2 | 6060 | 0.463 | 0.0249 | No | ||
| 42 | MPZ | 6245 | 0.429 | 0.0177 | No | ||
| 43 | SEMA4G | 6292 | 0.420 | 0.0179 | No | ||
| 44 | INSRR | 6363 | 0.408 | 0.0167 | No | ||
| 45 | ANK3 | 6454 | 0.391 | 0.0143 | No | ||
| 46 | NBL1 | 6592 | 0.371 | 0.0093 | No | ||
| 47 | LGI1 | 6646 | 0.364 | 0.0087 | No | ||
| 48 | HECTD2 | 6844 | 0.331 | 0.0002 | No | ||
| 49 | NCAM1 | 6965 | 0.310 | -0.0043 | No | ||
| 50 | HOXC8 | 7067 | 0.290 | -0.0079 | No | ||
| 51 | ASCL2 | 7237 | 0.261 | -0.0154 | No | ||
| 52 | CKAP4 | 7290 | 0.252 | -0.0166 | No | ||
| 53 | RFX4 | 7291 | 0.252 | -0.0150 | No | ||
| 54 | STMN1 | 7625 | 0.200 | -0.0317 | No | ||
| 55 | B3GALT2 | 8511 | 0.060 | -0.0792 | No | ||
| 56 | PNLIPRP2 | 8937 | -0.006 | -0.1022 | No | ||
| 57 | PALMD | 9115 | -0.034 | -0.1115 | No | ||
| 58 | SPON1 | 9350 | -0.070 | -0.1237 | No | ||
| 59 | FKBP2 | 9383 | -0.075 | -0.1250 | No | ||
| 60 | TPH2 | 9652 | -0.112 | -0.1388 | No | ||
| 61 | NRAS | 9973 | -0.163 | -0.1550 | No | ||
| 62 | EGR2 | 10935 | -0.314 | -0.2050 | No | ||
| 63 | EFNA1 | 10955 | -0.317 | -0.2040 | No | ||
| 64 | BAMBI | 10968 | -0.320 | -0.2026 | No | ||
| 65 | PAX6 | 11077 | -0.339 | -0.2063 | No | ||
| 66 | DPYSL4 | 11462 | -0.401 | -0.2245 | No | ||
| 67 | EDG1 | 11531 | -0.413 | -0.2255 | No | ||
| 68 | PPAP2B | 11817 | -0.458 | -0.2380 | No | ||
| 69 | SH3GLB1 | 11930 | -0.475 | -0.2411 | No | ||
| 70 | USF1 | 11995 | -0.485 | -0.2414 | No | ||
| 71 | PLXDC2 | 12037 | -0.492 | -0.2405 | No | ||
| 72 | DHRS3 | 12415 | -0.558 | -0.2573 | No | ||
| 73 | DDX6 | 12457 | -0.566 | -0.2559 | No | ||
| 74 | TBX5 | 12470 | -0.569 | -0.2530 | No | ||
| 75 | CLSTN1 | 12493 | -0.575 | -0.2505 | No | ||
| 76 | MXI1 | 12651 | -0.604 | -0.2551 | No | ||
| 77 | PDE6C | 12678 | -0.609 | -0.2526 | No | ||
| 78 | KCNA1 | 12956 | -0.659 | -0.2634 | No | ||
| 79 | GGPS1 | 13095 | -0.683 | -0.2665 | No | ||
| 80 | RAB6A | 13122 | -0.686 | -0.2636 | No | ||
| 81 | EHF | 13386 | -0.727 | -0.2732 | No | ||
| 82 | CPEB3 | 13778 | -0.800 | -0.2892 | No | ||
| 83 | PIK3C2A | 14015 | -0.847 | -0.2966 | No | ||
| 84 | LEMD1 | 14788 | -1.013 | -0.3319 | No | ||
| 85 | DFFB | 14923 | -1.042 | -0.3325 | Yes | ||
| 86 | RBP3 | 14975 | -1.052 | -0.3285 | Yes | ||
| 87 | ABLIM1 | 15096 | -1.081 | -0.3281 | Yes | ||
| 88 | DUSP6 | 15288 | -1.133 | -0.3312 | Yes | ||
| 89 | TEAD4 | 15391 | -1.159 | -0.3294 | Yes | ||
| 90 | LAMC2 | 15498 | -1.188 | -0.3275 | Yes | ||
| 91 | NOX4 | 15662 | -1.227 | -0.3285 | Yes | ||
| 92 | THBS3 | 15671 | -1.230 | -0.3211 | Yes | ||
| 93 | ZZZ3 | 15885 | -1.292 | -0.3244 | Yes | ||
| 94 | SLC38A4 | 16076 | -1.351 | -0.3260 | Yes | ||
| 95 | GAB2 | 16162 | -1.376 | -0.3219 | Yes | ||
| 96 | NTS | 16184 | -1.388 | -0.3142 | Yes | ||
| 97 | ELF5 | 16367 | -1.452 | -0.3147 | Yes | ||
| 98 | LBX1 | 16404 | -1.467 | -0.3073 | Yes | ||
| 99 | ELAVL4 | 16464 | -1.492 | -0.3010 | Yes | ||
| 100 | F11R | 16530 | -1.515 | -0.2949 | Yes | ||
| 101 | PRKAG1 | 16600 | -1.541 | -0.2888 | Yes | ||
| 102 | EIF4G2 | 16666 | -1.569 | -0.2823 | Yes | ||
| 103 | LHX9 | 16705 | -1.586 | -0.2742 | Yes | ||
| 104 | RBM8A | 16715 | -1.590 | -0.2646 | Yes | ||
| 105 | EFNA4 | 16739 | -1.599 | -0.2556 | Yes | ||
| 106 | GRK5 | 16830 | -1.642 | -0.2500 | Yes | ||
| 107 | EMX2 | 16919 | -1.681 | -0.2441 | Yes | ||
| 108 | DEDD | 16921 | -1.682 | -0.2334 | Yes | ||
| 109 | HABP2 | 17060 | -1.757 | -0.2297 | Yes | ||
| 110 | ESRRG | 17214 | -1.841 | -0.2262 | Yes | ||
| 111 | VDR | 17328 | -1.913 | -0.2201 | Yes | ||
| 112 | CSAD | 17360 | -1.927 | -0.2095 | Yes | ||
| 113 | PDE3B | 17406 | -1.953 | -0.1995 | Yes | ||
| 114 | EVA1 | 17622 | -2.123 | -0.1976 | Yes | ||
| 115 | SLC26A9 | 17927 | -2.414 | -0.1986 | Yes | ||
| 116 | LRP6 | 17949 | -2.447 | -0.1842 | Yes | ||
| 117 | RBM4 | 18038 | -2.566 | -0.1726 | Yes | ||
| 118 | HOXC4 | 18197 | -2.860 | -0.1629 | Yes | ||
| 119 | DDIT4 | 18270 | -3.044 | -0.1474 | Yes | ||
| 120 | PIK3CD | 18319 | -3.174 | -0.1297 | Yes | ||
| 121 | ING4 | 18363 | -3.343 | -0.1107 | Yes | ||
| 122 | MTX1 | 18464 | -3.877 | -0.0914 | Yes | ||
| 123 | CDKN2C | 18551 | -5.001 | -0.0642 | Yes | ||
| 124 | PDCD4 | 18553 | -5.062 | -0.0320 | Yes | ||
| 125 | GABARAPL1 | 18568 | -5.545 | 0.0026 | Yes |