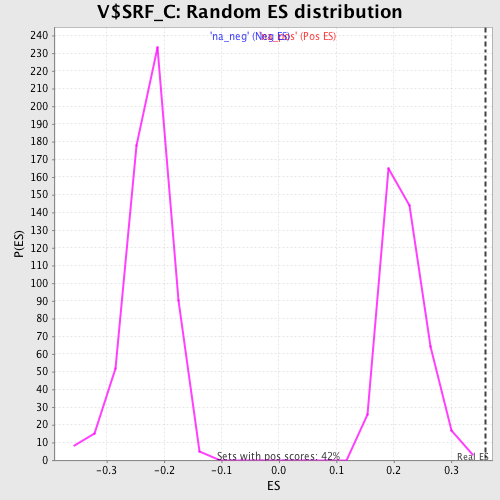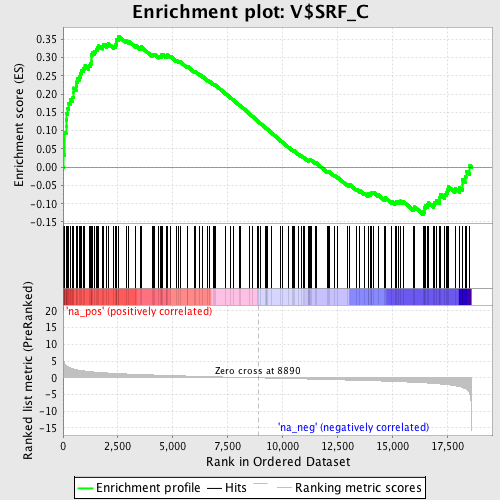
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$SRF_C |
| Enrichment Score (ES) | 0.35900512 |
| Normalized Enrichment Score (NES) | 1.6533035 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.23969312 |
| FWER p-Value | 0.63 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MYLK | 15 | 7.249 | 0.0353 | Yes | ||
| 2 | MRVI1 | 75 | 4.267 | 0.0534 | Yes | ||
| 3 | EML4 | 76 | 4.265 | 0.0746 | Yes | ||
| 4 | MYL9 | 77 | 4.222 | 0.0956 | Yes | ||
| 5 | CNN2 | 136 | 3.760 | 0.1112 | Yes | ||
| 6 | RASSF2 | 140 | 3.724 | 0.1296 | Yes | ||
| 7 | ASB2 | 151 | 3.660 | 0.1473 | Yes | ||
| 8 | CACNA1B | 208 | 3.332 | 0.1609 | Yes | ||
| 9 | VCL | 249 | 3.154 | 0.1744 | Yes | ||
| 10 | GADD45G | 319 | 2.953 | 0.1854 | Yes | ||
| 11 | FLNA | 445 | 2.691 | 0.1920 | Yes | ||
| 12 | TADA3L | 467 | 2.648 | 0.2041 | Yes | ||
| 13 | TANK | 473 | 2.638 | 0.2169 | Yes | ||
| 14 | SRF | 594 | 2.447 | 0.2226 | Yes | ||
| 15 | LCP1 | 618 | 2.419 | 0.2334 | Yes | ||
| 16 | SSH3 | 661 | 2.372 | 0.2430 | Yes | ||
| 17 | TCF2 | 746 | 2.254 | 0.2497 | Yes | ||
| 18 | MAPK14 | 804 | 2.181 | 0.2574 | Yes | ||
| 19 | SIPA1 | 847 | 2.148 | 0.2659 | Yes | ||
| 20 | PNKP | 931 | 2.063 | 0.2716 | Yes | ||
| 21 | SLC7A1 | 996 | 2.009 | 0.2782 | Yes | ||
| 22 | POPDC2 | 1179 | 1.878 | 0.2777 | Yes | ||
| 23 | TNNC1 | 1239 | 1.840 | 0.2836 | Yes | ||
| 24 | PHF12 | 1274 | 1.822 | 0.2909 | Yes | ||
| 25 | TLE3 | 1277 | 1.821 | 0.2998 | Yes | ||
| 26 | EMILIN2 | 1278 | 1.820 | 0.3089 | Yes | ||
| 27 | FOXP2 | 1326 | 1.787 | 0.3153 | Yes | ||
| 28 | CYR61 | 1430 | 1.718 | 0.3182 | Yes | ||
| 29 | CDC14A | 1498 | 1.677 | 0.3230 | Yes | ||
| 30 | MYH11 | 1554 | 1.650 | 0.3282 | Yes | ||
| 31 | TPM3 | 1628 | 1.613 | 0.3323 | Yes | ||
| 32 | RRM1 | 1788 | 1.537 | 0.3313 | Yes | ||
| 33 | MYL7 | 1818 | 1.527 | 0.3374 | Yes | ||
| 34 | LMCD1 | 1992 | 1.455 | 0.3352 | Yes | ||
| 35 | KCTD15 | 2058 | 1.425 | 0.3388 | Yes | ||
| 36 | LPP | 2315 | 1.331 | 0.3316 | Yes | ||
| 37 | GPR20 | 2375 | 1.314 | 0.3349 | Yes | ||
| 38 | MCAM | 2418 | 1.303 | 0.3391 | Yes | ||
| 39 | ERBB2IP | 2435 | 1.299 | 0.3447 | Yes | ||
| 40 | RGS3 | 2440 | 1.297 | 0.3510 | Yes | ||
| 41 | PDLIM4 | 2515 | 1.269 | 0.3533 | Yes | ||
| 42 | THBS1 | 2527 | 1.265 | 0.3590 | Yes | ||
| 43 | YWHAZ | 2875 | 1.159 | 0.3460 | No | ||
| 44 | IL17B | 2990 | 1.127 | 0.3454 | No | ||
| 45 | TGFB1I1 | 3305 | 1.043 | 0.3336 | No | ||
| 46 | CFL1 | 3509 | 0.989 | 0.3275 | No | ||
| 47 | DNAJB4 | 3552 | 0.975 | 0.3301 | No | ||
| 48 | COL1A2 | 4075 | 0.855 | 0.3061 | No | ||
| 49 | ROCK2 | 4096 | 0.851 | 0.3093 | No | ||
| 50 | ASPA | 4181 | 0.832 | 0.3089 | No | ||
| 51 | JPH2 | 4364 | 0.786 | 0.3029 | No | ||
| 52 | PPP1R12A | 4432 | 0.774 | 0.3031 | No | ||
| 53 | SPAG7 | 4468 | 0.763 | 0.3050 | No | ||
| 54 | NPAS2 | 4470 | 0.763 | 0.3088 | No | ||
| 55 | FOXP1 | 4533 | 0.750 | 0.3092 | No | ||
| 56 | MYL6 | 4709 | 0.715 | 0.3032 | No | ||
| 57 | TITF1 | 4727 | 0.710 | 0.3059 | No | ||
| 58 | MEIS1 | 4742 | 0.708 | 0.3086 | No | ||
| 59 | MUS81 | 4881 | 0.681 | 0.3045 | No | ||
| 60 | ACTG2 | 5150 | 0.629 | 0.2932 | No | ||
| 61 | DIXDC1 | 5262 | 0.609 | 0.2902 | No | ||
| 62 | MITF | 5336 | 0.594 | 0.2892 | No | ||
| 63 | USP52 | 5651 | 0.539 | 0.2749 | No | ||
| 64 | FHL2 | 5668 | 0.534 | 0.2767 | No | ||
| 65 | SRD5A2 | 6004 | 0.472 | 0.2609 | No | ||
| 66 | LRP5 | 6043 | 0.465 | 0.2611 | No | ||
| 67 | NKX2-2 | 6218 | 0.434 | 0.2539 | No | ||
| 68 | ITGA7 | 6354 | 0.409 | 0.2486 | No | ||
| 69 | STAT5B | 6594 | 0.371 | 0.2375 | No | ||
| 70 | RASD2 | 6677 | 0.358 | 0.2348 | No | ||
| 71 | FOSB | 6847 | 0.331 | 0.2273 | No | ||
| 72 | BCAS3 | 6913 | 0.319 | 0.2254 | No | ||
| 73 | HOXD10 | 6945 | 0.313 | 0.2253 | No | ||
| 74 | PADI2 | 7411 | 0.233 | 0.2012 | No | ||
| 75 | NR2F2 | 7629 | 0.200 | 0.1905 | No | ||
| 76 | HOXA5 | 7761 | 0.180 | 0.1843 | No | ||
| 77 | FOXE3 | 8044 | 0.131 | 0.1697 | No | ||
| 78 | STARD13 | 8080 | 0.125 | 0.1684 | No | ||
| 79 | DUSP2 | 8516 | 0.059 | 0.1451 | No | ||
| 80 | TAGLN | 8624 | 0.044 | 0.1395 | No | ||
| 81 | ACTR3 | 8860 | 0.006 | 0.1268 | No | ||
| 82 | MYO1E | 8890 | -0.000 | 0.1253 | No | ||
| 83 | SLC2A4 | 8997 | -0.015 | 0.1196 | No | ||
| 84 | PRKAB2 | 9231 | -0.051 | 0.1072 | No | ||
| 85 | PDLIM7 | 9259 | -0.055 | 0.1060 | No | ||
| 86 | PLN | 9263 | -0.056 | 0.1061 | No | ||
| 87 | AKAP12 | 9294 | -0.061 | 0.1048 | No | ||
| 88 | CNN1 | 9490 | -0.088 | 0.0947 | No | ||
| 89 | FOSL1 | 9921 | -0.155 | 0.0722 | No | ||
| 90 | EGR4 | 9994 | -0.166 | 0.0691 | No | ||
| 91 | KRTCAP2 | 10283 | -0.208 | 0.0545 | No | ||
| 92 | PLCB3 | 10467 | -0.234 | 0.0458 | No | ||
| 93 | HOXB4 | 10509 | -0.243 | 0.0448 | No | ||
| 94 | NR2F1 | 10524 | -0.246 | 0.0452 | No | ||
| 95 | NR4A1 | 10721 | -0.280 | 0.0360 | No | ||
| 96 | COL8A1 | 10844 | -0.299 | 0.0309 | No | ||
| 97 | EGR2 | 10935 | -0.314 | 0.0276 | No | ||
| 98 | ITGB1BP2 | 10996 | -0.325 | 0.0260 | No | ||
| 99 | JUNB | 11166 | -0.354 | 0.0186 | No | ||
| 100 | CRK | 11204 | -0.361 | 0.0184 | No | ||
| 101 | KCNMB1 | 11220 | -0.364 | 0.0194 | No | ||
| 102 | DUSP10 | 11245 | -0.368 | 0.0199 | No | ||
| 103 | LIN28 | 11267 | -0.372 | 0.0206 | No | ||
| 104 | IER2 | 11321 | -0.381 | 0.0196 | No | ||
| 105 | GFPT2 | 11514 | -0.410 | 0.0113 | No | ||
| 106 | POU3F4 | 11527 | -0.411 | 0.0127 | No | ||
| 107 | TUFT1 | 12038 | -0.493 | -0.0125 | No | ||
| 108 | HOXB5 | 12082 | -0.501 | -0.0123 | No | ||
| 109 | HOXA3 | 12126 | -0.508 | -0.0121 | No | ||
| 110 | MBNL1 | 12354 | -0.548 | -0.0217 | No | ||
| 111 | CALD1 | 12507 | -0.576 | -0.0270 | No | ||
| 112 | ARPC4 | 12946 | -0.657 | -0.0475 | No | ||
| 113 | RUNDC1 | 13047 | -0.677 | -0.0495 | No | ||
| 114 | MYO1B | 13056 | -0.678 | -0.0466 | No | ||
| 115 | DLL1 | 13383 | -0.726 | -0.0606 | No | ||
| 116 | ITGB6 | 13525 | -0.756 | -0.0645 | No | ||
| 117 | TAZ | 13726 | -0.792 | -0.0714 | No | ||
| 118 | EGR1 | 13899 | -0.822 | -0.0766 | No | ||
| 119 | PFN1 | 13903 | -0.822 | -0.0727 | No | ||
| 120 | SYNCRIP | 13996 | -0.841 | -0.0735 | No | ||
| 121 | DVL3 | 14034 | -0.849 | -0.0713 | No | ||
| 122 | AOC3 | 14071 | -0.856 | -0.0689 | No | ||
| 123 | TRIM46 | 14145 | -0.872 | -0.0686 | No | ||
| 124 | MRGPRF | 14354 | -0.915 | -0.0753 | No | ||
| 125 | DAPK3 | 14639 | -0.978 | -0.0858 | No | ||
| 126 | SLC25A4 | 14691 | -0.987 | -0.0836 | No | ||
| 127 | CKM | 14977 | -1.053 | -0.0938 | No | ||
| 128 | SCOC | 15127 | -1.089 | -0.0965 | No | ||
| 129 | GGN | 15191 | -1.107 | -0.0944 | No | ||
| 130 | DUSP6 | 15288 | -1.133 | -0.0939 | No | ||
| 131 | DSTN | 15366 | -1.153 | -0.0924 | No | ||
| 132 | FGFRL1 | 15512 | -1.191 | -0.0943 | No | ||
| 133 | MAP1A | 15978 | -1.319 | -0.1129 | No | ||
| 134 | TOP1 | 16024 | -1.334 | -0.1087 | No | ||
| 135 | NKX6-2 | 16408 | -1.467 | -0.1221 | No | ||
| 136 | ELAVL4 | 16464 | -1.492 | -0.1177 | No | ||
| 137 | MATR3 | 16465 | -1.492 | -0.1102 | No | ||
| 138 | EGR3 | 16523 | -1.514 | -0.1058 | No | ||
| 139 | GRK6 | 16593 | -1.539 | -0.1019 | No | ||
| 140 | CAP1 | 16637 | -1.558 | -0.0964 | No | ||
| 141 | FOS | 16885 | -1.668 | -0.1015 | No | ||
| 142 | RSU1 | 16935 | -1.688 | -0.0957 | No | ||
| 143 | KCNK3 | 16997 | -1.726 | -0.0904 | No | ||
| 144 | ANXA6 | 17166 | -1.812 | -0.0905 | No | ||
| 145 | MYO18B | 17171 | -1.814 | -0.0817 | No | ||
| 146 | EFHD1 | 17208 | -1.838 | -0.0745 | No | ||
| 147 | RHOJ | 17404 | -1.952 | -0.0753 | No | ||
| 148 | ABL1 | 17458 | -1.997 | -0.0683 | No | ||
| 149 | ACTA1 | 17524 | -2.040 | -0.0616 | No | ||
| 150 | LTBP1 | 17570 | -2.081 | -0.0537 | No | ||
| 151 | CFL2 | 17862 | -2.341 | -0.0578 | No | ||
| 152 | ACTB | 18073 | -2.633 | -0.0560 | No | ||
| 153 | ARHGAP1 | 18198 | -2.863 | -0.0485 | No | ||
| 154 | ACVR1 | 18200 | -2.870 | -0.0343 | No | ||
| 155 | DMD | 18322 | -3.185 | -0.0249 | No | ||
| 156 | RBBP7 | 18365 | -3.348 | -0.0105 | No | ||
| 157 | CA3 | 18542 | -4.835 | 0.0040 | No |