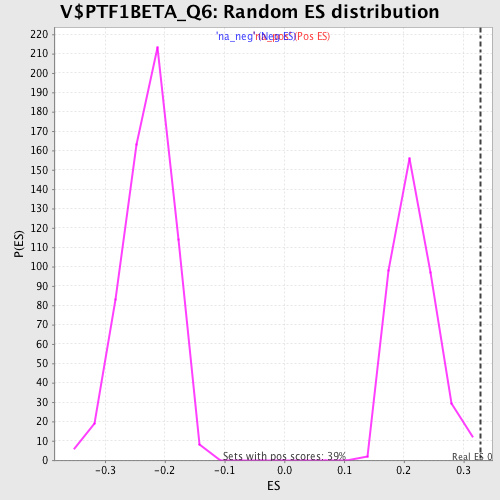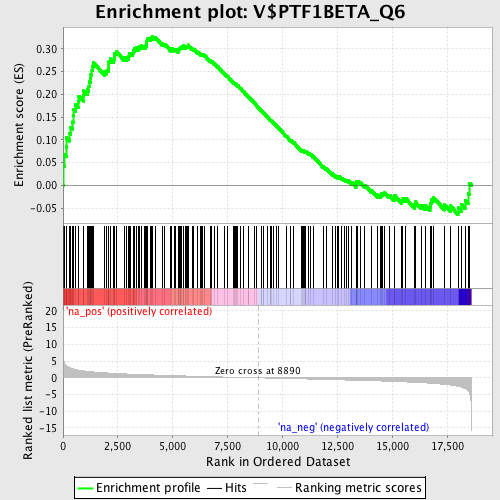
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$PTF1BETA_Q6 |
| Enrichment Score (ES) | 0.32765332 |
| Normalized Enrichment Score (NES) | 1.5027581 |
| Nominal p-value | 0.007614213 |
| FDR q-value | 0.24708712 |
| FWER p-Value | 0.981 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EMP3 | 12 | 7.482 | 0.0426 | Yes | ||
| 2 | LPL | 47 | 4.744 | 0.0682 | Yes | ||
| 3 | RASSF2 | 140 | 3.724 | 0.0847 | Yes | ||
| 4 | RELB | 157 | 3.603 | 0.1047 | Yes | ||
| 5 | STAT5A | 291 | 3.017 | 0.1149 | Yes | ||
| 6 | ARF3 | 350 | 2.862 | 0.1283 | Yes | ||
| 7 | ELA1 | 437 | 2.707 | 0.1393 | Yes | ||
| 8 | KLF13 | 459 | 2.658 | 0.1535 | Yes | ||
| 9 | DNMT3A | 478 | 2.630 | 0.1677 | Yes | ||
| 10 | FCHSD2 | 568 | 2.488 | 0.1773 | Yes | ||
| 11 | TNFRSF1A | 694 | 2.313 | 0.1839 | Yes | ||
| 12 | C20ORF185 | 712 | 2.290 | 0.1962 | Yes | ||
| 13 | ROBO1 | 917 | 2.075 | 0.1971 | Yes | ||
| 14 | HOXA11 | 940 | 2.059 | 0.2078 | Yes | ||
| 15 | ZFP36L1 | 1112 | 1.921 | 0.2097 | Yes | ||
| 16 | CHD2 | 1173 | 1.882 | 0.2173 | Yes | ||
| 17 | THRA | 1188 | 1.875 | 0.2274 | Yes | ||
| 18 | SRGAP2 | 1235 | 1.841 | 0.2355 | Yes | ||
| 19 | H1F0 | 1266 | 1.824 | 0.2444 | Yes | ||
| 20 | APBA1 | 1314 | 1.799 | 0.2523 | Yes | ||
| 21 | FOXP2 | 1326 | 1.787 | 0.2620 | Yes | ||
| 22 | GATA6 | 1368 | 1.755 | 0.2699 | Yes | ||
| 23 | KCNJ1 | 1887 | 1.493 | 0.2505 | Yes | ||
| 24 | UBTF | 1999 | 1.452 | 0.2529 | Yes | ||
| 25 | KCTD15 | 2058 | 1.425 | 0.2580 | Yes | ||
| 26 | ETV6 | 2086 | 1.415 | 0.2647 | Yes | ||
| 27 | ZIC1 | 2089 | 1.415 | 0.2728 | Yes | ||
| 28 | CRHR1 | 2149 | 1.397 | 0.2776 | Yes | ||
| 29 | IMPDH1 | 2303 | 1.334 | 0.2771 | Yes | ||
| 30 | PTPN2 | 2339 | 1.324 | 0.2828 | Yes | ||
| 31 | HOXD12 | 2348 | 1.321 | 0.2900 | Yes | ||
| 32 | HMGB3 | 2415 | 1.303 | 0.2940 | Yes | ||
| 33 | FGF14 | 2778 | 1.189 | 0.2812 | Yes | ||
| 34 | SLC25A14 | 2907 | 1.150 | 0.2809 | Yes | ||
| 35 | TRERF1 | 2988 | 1.128 | 0.2831 | Yes | ||
| 36 | ADCK4 | 3006 | 1.122 | 0.2887 | Yes | ||
| 37 | ACCN3 | 3092 | 1.098 | 0.2904 | Yes | ||
| 38 | DUSP22 | 3185 | 1.076 | 0.2916 | Yes | ||
| 39 | TRAF4 | 3217 | 1.066 | 0.2961 | Yes | ||
| 40 | BCL2 | 3260 | 1.054 | 0.2999 | Yes | ||
| 41 | PAX3 | 3325 | 1.039 | 0.3025 | Yes | ||
| 42 | SLITRK3 | 3439 | 1.008 | 0.3022 | Yes | ||
| 43 | XPO1 | 3496 | 0.991 | 0.3049 | Yes | ||
| 44 | ANKRD17 | 3573 | 0.969 | 0.3064 | Yes | ||
| 45 | FBXO40 | 3693 | 0.941 | 0.3054 | Yes | ||
| 46 | PIAS2 | 3775 | 0.921 | 0.3063 | Yes | ||
| 47 | THRAP1 | 3795 | 0.918 | 0.3106 | Yes | ||
| 48 | CAPG | 3814 | 0.914 | 0.3149 | Yes | ||
| 49 | CDH13 | 3823 | 0.911 | 0.3197 | Yes | ||
| 50 | REST | 3865 | 0.901 | 0.3227 | Yes | ||
| 51 | NXPH3 | 3972 | 0.881 | 0.3220 | Yes | ||
| 52 | BCL2L1 | 4023 | 0.869 | 0.3244 | Yes | ||
| 53 | SLC25A13 | 4055 | 0.861 | 0.3277 | Yes | ||
| 54 | SYT7 | 4205 | 0.826 | 0.3244 | No | ||
| 55 | PMP22 | 4537 | 0.749 | 0.3107 | No | ||
| 56 | NME3 | 4626 | 0.731 | 0.3102 | No | ||
| 57 | BBOX1 | 4913 | 0.676 | 0.2986 | No | ||
| 58 | LRRTM4 | 4945 | 0.670 | 0.3008 | No | ||
| 59 | SOX4 | 5055 | 0.649 | 0.2986 | No | ||
| 60 | GNG3 | 5123 | 0.634 | 0.2987 | No | ||
| 61 | JMJD1B | 5240 | 0.612 | 0.2959 | No | ||
| 62 | NR4A3 | 5261 | 0.609 | 0.2984 | No | ||
| 63 | CCKBR | 5284 | 0.605 | 0.3007 | No | ||
| 64 | LIF | 5329 | 0.596 | 0.3017 | No | ||
| 65 | NTRK1 | 5367 | 0.589 | 0.3031 | No | ||
| 66 | CASK | 5415 | 0.581 | 0.3039 | No | ||
| 67 | EED | 5476 | 0.569 | 0.3040 | No | ||
| 68 | RIMS1 | 5485 | 0.567 | 0.3068 | No | ||
| 69 | BAD | 5594 | 0.547 | 0.3041 | No | ||
| 70 | FBXO24 | 5637 | 0.540 | 0.3050 | No | ||
| 71 | EIF4G1 | 5685 | 0.531 | 0.3055 | No | ||
| 72 | GABRA1 | 5692 | 0.530 | 0.3082 | No | ||
| 73 | TEAD1 | 5892 | 0.491 | 0.3003 | No | ||
| 74 | EEF1A2 | 5962 | 0.481 | 0.2993 | No | ||
| 75 | ITSN2 | 6130 | 0.450 | 0.2929 | No | ||
| 76 | REPIN1 | 6259 | 0.427 | 0.2884 | No | ||
| 77 | CTCF | 6306 | 0.417 | 0.2883 | No | ||
| 78 | ITGA7 | 6354 | 0.409 | 0.2882 | No | ||
| 79 | ANK3 | 6454 | 0.391 | 0.2851 | No | ||
| 80 | OBSCN | 6708 | 0.355 | 0.2734 | No | ||
| 81 | STC1 | 6780 | 0.343 | 0.2715 | No | ||
| 82 | ITGB7 | 6905 | 0.320 | 0.2667 | No | ||
| 83 | SLC6A14 | 7042 | 0.295 | 0.2610 | No | ||
| 84 | SMOC1 | 7346 | 0.242 | 0.2460 | No | ||
| 85 | BSCL2 | 7485 | 0.221 | 0.2398 | No | ||
| 86 | ERBB3 | 7776 | 0.177 | 0.2251 | No | ||
| 87 | PLEKHC1 | 7817 | 0.170 | 0.2239 | No | ||
| 88 | NPPA | 7834 | 0.168 | 0.2240 | No | ||
| 89 | RASA1 | 7892 | 0.159 | 0.2218 | No | ||
| 90 | PANK3 | 7970 | 0.144 | 0.2185 | No | ||
| 91 | STARD13 | 8080 | 0.125 | 0.2133 | No | ||
| 92 | PDE10A | 8223 | 0.106 | 0.2062 | No | ||
| 93 | SMAD6 | 8431 | 0.074 | 0.1954 | No | ||
| 94 | HES7 | 8470 | 0.068 | 0.1938 | No | ||
| 95 | CD163 | 8721 | 0.029 | 0.1804 | No | ||
| 96 | PLXNC1 | 8835 | 0.011 | 0.1743 | No | ||
| 97 | MATN1 | 9054 | -0.024 | 0.1626 | No | ||
| 98 | VAX1 | 9061 | -0.025 | 0.1625 | No | ||
| 99 | PAQR4 | 9149 | -0.039 | 0.1580 | No | ||
| 100 | PTBP2 | 9151 | -0.039 | 0.1582 | No | ||
| 101 | TREX2 | 9319 | -0.065 | 0.1495 | No | ||
| 102 | USP5 | 9438 | -0.082 | 0.1436 | No | ||
| 103 | KCNE1L | 9465 | -0.085 | 0.1426 | No | ||
| 104 | SLITRK5 | 9493 | -0.088 | 0.1417 | No | ||
| 105 | TNIP2 | 9601 | -0.105 | 0.1365 | No | ||
| 106 | LRRTM1 | 9740 | -0.125 | 0.1297 | No | ||
| 107 | MDS1 | 9832 | -0.140 | 0.1256 | No | ||
| 108 | CCNC | 10185 | -0.193 | 0.1077 | No | ||
| 109 | YWHAE | 10376 | -0.220 | 0.0986 | No | ||
| 110 | TBL1X | 10384 | -0.221 | 0.0995 | No | ||
| 111 | HOXB4 | 10509 | -0.243 | 0.0942 | No | ||
| 112 | PUM1 | 10867 | -0.302 | 0.0766 | No | ||
| 113 | MAN2C1 | 10894 | -0.307 | 0.0770 | No | ||
| 114 | ZFYVE20 | 10941 | -0.315 | 0.0763 | No | ||
| 115 | NPDC1 | 11016 | -0.328 | 0.0742 | No | ||
| 116 | SYT3 | 11041 | -0.332 | 0.0748 | No | ||
| 117 | GNAT1 | 11065 | -0.337 | 0.0755 | No | ||
| 118 | CRK | 11204 | -0.361 | 0.0701 | No | ||
| 119 | LIN28 | 11267 | -0.372 | 0.0689 | No | ||
| 120 | AMMECR1 | 11420 | -0.395 | 0.0630 | No | ||
| 121 | NR4A2 | 11857 | -0.463 | 0.0420 | No | ||
| 122 | HOXB6 | 12000 | -0.486 | 0.0371 | No | ||
| 123 | IGFBP6 | 12271 | -0.532 | 0.0256 | No | ||
| 124 | DHRS3 | 12415 | -0.558 | 0.0211 | No | ||
| 125 | CALD1 | 12507 | -0.576 | 0.0195 | No | ||
| 126 | TBCC | 12573 | -0.590 | 0.0193 | No | ||
| 127 | NCALD | 12694 | -0.612 | 0.0164 | No | ||
| 128 | TMPRSS2 | 12812 | -0.631 | 0.0137 | No | ||
| 129 | AR | 12918 | -0.651 | 0.0118 | No | ||
| 130 | ARMCX3 | 13007 | -0.669 | 0.0109 | No | ||
| 131 | DICER1 | 13161 | -0.693 | 0.0066 | No | ||
| 132 | ITPKC | 13350 | -0.721 | 0.0005 | No | ||
| 133 | PHYHIP | 13359 | -0.723 | 0.0043 | No | ||
| 134 | ROR1 | 13363 | -0.723 | 0.0083 | No | ||
| 135 | ATP1B3 | 13418 | -0.732 | 0.0096 | No | ||
| 136 | MTMR4 | 13555 | -0.761 | 0.0066 | No | ||
| 137 | HHIP | 13754 | -0.796 | 0.0005 | No | ||
| 138 | LRCH4 | 14054 | -0.853 | -0.0108 | No | ||
| 139 | CD6 | 14336 | -0.912 | -0.0207 | No | ||
| 140 | WISP1 | 14458 | -0.935 | -0.0219 | No | ||
| 141 | PCDH7 | 14496 | -0.944 | -0.0184 | No | ||
| 142 | FASTK | 14570 | -0.961 | -0.0168 | No | ||
| 143 | SLC8A3 | 14645 | -0.978 | -0.0152 | No | ||
| 144 | PPARGC1A | 14863 | -1.028 | -0.0210 | No | ||
| 145 | PPP2R2A | 15090 | -1.079 | -0.0270 | No | ||
| 146 | XPR1 | 15107 | -1.085 | -0.0216 | No | ||
| 147 | KIRREL2 | 15424 | -1.167 | -0.0320 | No | ||
| 148 | TGFB3 | 15487 | -1.185 | -0.0285 | No | ||
| 149 | CITED1 | 15605 | -1.214 | -0.0278 | No | ||
| 150 | ST18 | 16018 | -1.333 | -0.0424 | No | ||
| 151 | PURA | 16040 | -1.338 | -0.0358 | No | ||
| 152 | GPC6 | 16331 | -1.440 | -0.0432 | No | ||
| 153 | GSPT1 | 16507 | -1.506 | -0.0440 | No | ||
| 154 | STOML2 | 16722 | -1.592 | -0.0464 | No | ||
| 155 | CDK6 | 16749 | -1.603 | -0.0385 | No | ||
| 156 | SNCAIP | 16787 | -1.620 | -0.0312 | No | ||
| 157 | NMT1 | 16871 | -1.659 | -0.0261 | No | ||
| 158 | ZIC3 | 17383 | -1.941 | -0.0426 | No | ||
| 159 | ACADVL | 17639 | -2.134 | -0.0441 | No | ||
| 160 | LRRFIP2 | 18001 | -2.528 | -0.0490 | No | ||
| 161 | CSNK1E | 18143 | -2.747 | -0.0408 | No | ||
| 162 | DMD | 18322 | -3.185 | -0.0320 | No | ||
| 163 | MAGED1 | 18458 | -3.802 | -0.0174 | No | ||
| 164 | HEBP2 | 18519 | -4.475 | 0.0053 | No |