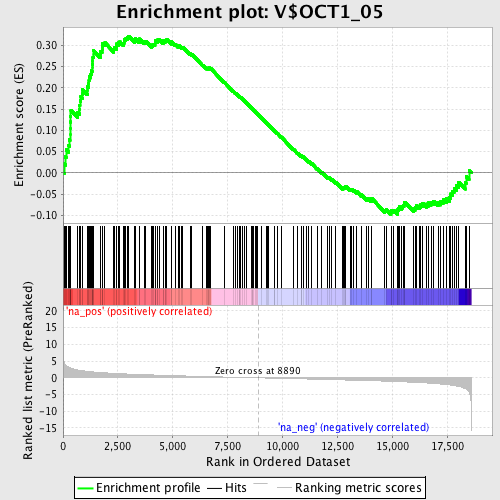
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$OCT1_05 |
| Enrichment Score (ES) | 0.32165873 |
| Normalized Enrichment Score (NES) | 1.4807565 |
| Nominal p-value | 0.004842615 |
| FDR q-value | 0.2622094 |
| FWER p-Value | 0.989 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | LPL | 47 | 4.744 | 0.0211 | Yes | ||
| 2 | SCNN1A | 104 | 3.978 | 0.0379 | Yes | ||
| 3 | DGKG | 139 | 3.731 | 0.0547 | Yes | ||
| 4 | STAT4 | 253 | 3.142 | 0.0642 | Yes | ||
| 5 | TBXAS1 | 281 | 3.054 | 0.0780 | Yes | ||
| 6 | ARHGAP4 | 315 | 2.964 | 0.0910 | Yes | ||
| 7 | GADD45G | 319 | 2.953 | 0.1056 | Yes | ||
| 8 | LIG4 | 325 | 2.931 | 0.1199 | Yes | ||
| 9 | NEDD9 | 336 | 2.890 | 0.1338 | Yes | ||
| 10 | POU2F1 | 356 | 2.857 | 0.1470 | Yes | ||
| 11 | CDK2 | 668 | 2.362 | 0.1419 | Yes | ||
| 12 | TCF2 | 746 | 2.254 | 0.1490 | Yes | ||
| 13 | ITPR3 | 759 | 2.234 | 0.1595 | Yes | ||
| 14 | RRAS | 770 | 2.216 | 0.1700 | Yes | ||
| 15 | GCM1 | 807 | 2.178 | 0.1789 | Yes | ||
| 16 | HIVEP3 | 873 | 2.122 | 0.1860 | Yes | ||
| 17 | NIN | 889 | 2.100 | 0.1956 | Yes | ||
| 18 | ZFP36L1 | 1112 | 1.921 | 0.1932 | Yes | ||
| 19 | PRRX1 | 1117 | 1.920 | 0.2025 | Yes | ||
| 20 | PROM1 | 1163 | 1.890 | 0.2095 | Yes | ||
| 21 | GSC | 1177 | 1.881 | 0.2182 | Yes | ||
| 22 | THRA | 1188 | 1.875 | 0.2270 | Yes | ||
| 23 | ING1 | 1261 | 1.827 | 0.2322 | Yes | ||
| 24 | ATP2A2 | 1283 | 1.815 | 0.2401 | Yes | ||
| 25 | PRKCB1 | 1323 | 1.788 | 0.2469 | Yes | ||
| 26 | FOXP2 | 1326 | 1.787 | 0.2558 | Yes | ||
| 27 | SVIL | 1350 | 1.766 | 0.2633 | Yes | ||
| 28 | SATB2 | 1358 | 1.762 | 0.2717 | Yes | ||
| 29 | NRP2 | 1381 | 1.748 | 0.2793 | Yes | ||
| 30 | IMPG2 | 1382 | 1.748 | 0.2880 | Yes | ||
| 31 | SOX5 | 1703 | 1.576 | 0.2785 | Yes | ||
| 32 | EIF3S1 | 1710 | 1.574 | 0.2860 | Yes | ||
| 33 | IRX2 | 1775 | 1.543 | 0.2902 | Yes | ||
| 34 | MTUS1 | 1783 | 1.539 | 0.2975 | Yes | ||
| 35 | SH3GL3 | 1797 | 1.534 | 0.3045 | Yes | ||
| 36 | LHX6 | 1907 | 1.486 | 0.3060 | Yes | ||
| 37 | LPP | 2315 | 1.331 | 0.2906 | Yes | ||
| 38 | PTPN2 | 2339 | 1.324 | 0.2959 | Yes | ||
| 39 | RGS3 | 2440 | 1.297 | 0.2970 | Yes | ||
| 40 | SYT1 | 2444 | 1.296 | 0.3033 | Yes | ||
| 41 | DCN | 2512 | 1.270 | 0.3060 | Yes | ||
| 42 | SMPX | 2588 | 1.246 | 0.3081 | Yes | ||
| 43 | BNC2 | 2759 | 1.193 | 0.3048 | Yes | ||
| 44 | FGF14 | 2778 | 1.189 | 0.3098 | Yes | ||
| 45 | BTK | 2804 | 1.181 | 0.3143 | Yes | ||
| 46 | PIK4CB | 2861 | 1.163 | 0.3171 | Yes | ||
| 47 | GNA14 | 2923 | 1.146 | 0.3195 | Yes | ||
| 48 | TRERF1 | 2988 | 1.128 | 0.3217 | Yes | ||
| 49 | BCL2 | 3260 | 1.054 | 0.3122 | No | ||
| 50 | HOXA10 | 3278 | 1.050 | 0.3165 | No | ||
| 51 | UBE3A | 3471 | 0.998 | 0.3111 | No | ||
| 52 | FSTL5 | 3489 | 0.993 | 0.3151 | No | ||
| 53 | VPREB3 | 3686 | 0.943 | 0.3092 | No | ||
| 54 | CPA4 | 3761 | 0.925 | 0.3098 | No | ||
| 55 | MID1 | 4032 | 0.866 | 0.2995 | No | ||
| 56 | HOXB9 | 4081 | 0.854 | 0.3012 | No | ||
| 57 | ITCH | 4124 | 0.846 | 0.3031 | No | ||
| 58 | COL12A1 | 4189 | 0.830 | 0.3038 | No | ||
| 59 | ENPP1 | 4191 | 0.829 | 0.3078 | No | ||
| 60 | HOXB3 | 4192 | 0.829 | 0.3120 | No | ||
| 61 | STMN2 | 4310 | 0.795 | 0.3096 | No | ||
| 62 | HIST2H2AC | 4321 | 0.794 | 0.3130 | No | ||
| 63 | ARMET | 4393 | 0.781 | 0.3131 | No | ||
| 64 | BHLHB5 | 4559 | 0.747 | 0.3078 | No | ||
| 65 | SCML4 | 4573 | 0.742 | 0.3108 | No | ||
| 66 | LPHN1 | 4647 | 0.727 | 0.3105 | No | ||
| 67 | DPYSL3 | 4684 | 0.719 | 0.3121 | No | ||
| 68 | TITF1 | 4727 | 0.710 | 0.3134 | No | ||
| 69 | SLC7A11 | 4928 | 0.673 | 0.3059 | No | ||
| 70 | PIM2 | 4930 | 0.673 | 0.3092 | No | ||
| 71 | SP6 | 5104 | 0.637 | 0.3030 | No | ||
| 72 | NR4A3 | 5261 | 0.609 | 0.2976 | No | ||
| 73 | HOXD11 | 5283 | 0.606 | 0.2995 | No | ||
| 74 | SERTAD4 | 5403 | 0.582 | 0.2959 | No | ||
| 75 | SFRS14 | 5462 | 0.571 | 0.2956 | No | ||
| 76 | RPP21 | 5783 | 0.512 | 0.2808 | No | ||
| 77 | SLC6A15 | 5864 | 0.495 | 0.2790 | No | ||
| 78 | MRAS | 6372 | 0.406 | 0.2535 | No | ||
| 79 | NRL | 6545 | 0.377 | 0.2461 | No | ||
| 80 | TSGA14 | 6569 | 0.374 | 0.2467 | No | ||
| 81 | PHOX2B | 6634 | 0.365 | 0.2450 | No | ||
| 82 | LGI1 | 6646 | 0.364 | 0.2463 | No | ||
| 83 | ACYP2 | 6659 | 0.362 | 0.2474 | No | ||
| 84 | TBR1 | 6716 | 0.353 | 0.2461 | No | ||
| 85 | YWHAB | 7343 | 0.243 | 0.2134 | No | ||
| 86 | HOXA5 | 7761 | 0.180 | 0.1917 | No | ||
| 87 | SILV | 7856 | 0.164 | 0.1874 | No | ||
| 88 | PANK4 | 7935 | 0.149 | 0.1839 | No | ||
| 89 | NEUROG1 | 8036 | 0.132 | 0.1792 | No | ||
| 90 | CENTD1 | 8057 | 0.129 | 0.1787 | No | ||
| 91 | HOXC5 | 8083 | 0.125 | 0.1780 | No | ||
| 92 | RGS13 | 8172 | 0.113 | 0.1738 | No | ||
| 93 | NR2E1 | 8273 | 0.099 | 0.1689 | No | ||
| 94 | HIST3H2A | 8379 | 0.083 | 0.1636 | No | ||
| 95 | ISL1 | 8576 | 0.051 | 0.1532 | No | ||
| 96 | LYN | 8625 | 0.044 | 0.1508 | No | ||
| 97 | IL6 | 8650 | 0.040 | 0.1497 | No | ||
| 98 | PTEN | 8691 | 0.035 | 0.1477 | No | ||
| 99 | TLL2 | 8785 | 0.019 | 0.1428 | No | ||
| 100 | RHOBTB3 | 8803 | 0.016 | 0.1419 | No | ||
| 101 | ADORA1 | 8863 | 0.005 | 0.1388 | No | ||
| 102 | FZD4 | 9029 | -0.021 | 0.1299 | No | ||
| 103 | SLC9A7 | 9284 | -0.059 | 0.1164 | No | ||
| 104 | TREX2 | 9319 | -0.065 | 0.1149 | No | ||
| 105 | HIST3H2BB | 9349 | -0.070 | 0.1137 | No | ||
| 106 | CDX1 | 9616 | -0.106 | 0.0998 | No | ||
| 107 | CRH | 9655 | -0.113 | 0.0983 | No | ||
| 108 | CDX4 | 9769 | -0.130 | 0.0928 | No | ||
| 109 | CPNE1 | 9938 | -0.158 | 0.0845 | No | ||
| 110 | EDN1 | 10501 | -0.241 | 0.0552 | No | ||
| 111 | LDB2 | 10687 | -0.275 | 0.0466 | No | ||
| 112 | CNNM4 | 10853 | -0.300 | 0.0391 | No | ||
| 113 | HMGB1 | 10861 | -0.301 | 0.0403 | No | ||
| 114 | EGR2 | 10935 | -0.314 | 0.0379 | No | ||
| 115 | PAX6 | 11077 | -0.339 | 0.0319 | No | ||
| 116 | ZHX2 | 11202 | -0.361 | 0.0270 | No | ||
| 117 | PCDH8 | 11333 | -0.382 | 0.0218 | No | ||
| 118 | NUDT3 | 11577 | -0.420 | 0.0108 | No | ||
| 119 | LRFN5 | 11798 | -0.455 | 0.0011 | No | ||
| 120 | RHOB | 12065 | -0.497 | -0.0108 | No | ||
| 121 | HOXA3 | 12126 | -0.508 | -0.0116 | No | ||
| 122 | SLC19A3 | 12254 | -0.530 | -0.0158 | No | ||
| 123 | GPRC5B | 12414 | -0.558 | -0.0216 | No | ||
| 124 | LRRN3 | 12746 | -0.620 | -0.0365 | No | ||
| 125 | SLIT3 | 12773 | -0.624 | -0.0348 | No | ||
| 126 | CDX2 | 12818 | -0.632 | -0.0340 | No | ||
| 127 | MAP2K6 | 12872 | -0.641 | -0.0337 | No | ||
| 128 | FGFR2 | 12882 | -0.642 | -0.0310 | No | ||
| 129 | AMBN | 13079 | -0.680 | -0.0382 | No | ||
| 130 | RARB | 13154 | -0.691 | -0.0388 | No | ||
| 131 | SOST | 13256 | -0.705 | -0.0407 | No | ||
| 132 | EHF | 13386 | -0.727 | -0.0441 | No | ||
| 133 | SEMA6C | 13593 | -0.768 | -0.0514 | No | ||
| 134 | POU2F3 | 13837 | -0.811 | -0.0606 | No | ||
| 135 | GPM6A | 13920 | -0.826 | -0.0609 | No | ||
| 136 | RHOBTB1 | 14057 | -0.854 | -0.0640 | No | ||
| 137 | GABRB1 | 14065 | -0.855 | -0.0601 | No | ||
| 138 | ASCL3 | 14668 | -0.982 | -0.0879 | No | ||
| 139 | LRRN1 | 14719 | -0.994 | -0.0856 | No | ||
| 140 | ELK3 | 14945 | -1.045 | -0.0926 | No | ||
| 141 | HOXB2 | 14962 | -1.049 | -0.0882 | No | ||
| 142 | EMILIN3 | 15061 | -1.073 | -0.0882 | No | ||
| 143 | PIM1 | 15228 | -1.119 | -0.0916 | No | ||
| 144 | FGF20 | 15247 | -1.124 | -0.0870 | No | ||
| 145 | DUSP6 | 15288 | -1.133 | -0.0835 | No | ||
| 146 | GALNT1 | 15309 | -1.137 | -0.0789 | No | ||
| 147 | TMOD2 | 15435 | -1.171 | -0.0799 | No | ||
| 148 | SIX1 | 15491 | -1.187 | -0.0769 | No | ||
| 149 | DKK1 | 15538 | -1.200 | -0.0734 | No | ||
| 150 | COL25A1 | 15564 | -1.207 | -0.0688 | No | ||
| 151 | TOPBP1 | 15980 | -1.319 | -0.0847 | No | ||
| 152 | PURA | 16040 | -1.338 | -0.0812 | No | ||
| 153 | NOS1 | 16090 | -1.357 | -0.0771 | No | ||
| 154 | TRPM1 | 16233 | -1.405 | -0.0778 | No | ||
| 155 | ALDH5A1 | 16301 | -1.429 | -0.0743 | No | ||
| 156 | CDCA7 | 16385 | -1.459 | -0.0715 | No | ||
| 157 | NFYB | 16556 | -1.524 | -0.0731 | No | ||
| 158 | HOXB1 | 16649 | -1.561 | -0.0703 | No | ||
| 159 | KCNN3 | 16768 | -1.611 | -0.0687 | No | ||
| 160 | FOS | 16885 | -1.668 | -0.0667 | No | ||
| 161 | RNF38 | 17092 | -1.776 | -0.0690 | No | ||
| 162 | ESRRG | 17214 | -1.841 | -0.0664 | No | ||
| 163 | HIST1H2AC | 17318 | -1.907 | -0.0624 | No | ||
| 164 | PKNOX2 | 17468 | -2.004 | -0.0605 | No | ||
| 165 | DLX1 | 17598 | -2.107 | -0.0570 | No | ||
| 166 | IGJ | 17659 | -2.145 | -0.0496 | No | ||
| 167 | DTNA | 17737 | -2.209 | -0.0427 | No | ||
| 168 | LECT1 | 17818 | -2.287 | -0.0357 | No | ||
| 169 | PDE4D | 17934 | -2.425 | -0.0298 | No | ||
| 170 | SEMA7A | 18015 | -2.541 | -0.0215 | No | ||
| 171 | DMD | 18322 | -3.185 | -0.0222 | No | ||
| 172 | TCF12 | 18369 | -3.363 | -0.0079 | No | ||
| 173 | GRIA3 | 18502 | -4.254 | 0.0062 | No |
